Glossary of cellular and molecular biology (0–L)
Appearance
(Redirected from Over expression)
This glossary of cellular and molecular biology is a list of definitions of terms and concepts commonly used in the study of cell biology, molecular biology, and related disciplines, including genetics, biochemistry, and microbiology.[1] It is split across two articles:
- This page, Glossary of cellular and molecular biology (0–L), lists terms beginning with numbers and with the letters A through L.
- Glossary of cellular and molecular biology (M–Z) lists terms beginning with the letters M through Z.
This glossary is intended as introductory material for novices (for more specific and technical detail, see the article corresponding to each term). It has been designed as a companion to Glossary of genetics and evolutionary biology, which contains many overlapping and related terms; other related glossaries include Glossary of virology and Glossary of chemistry.
0–9
[edit]- 3' untranslated region (3'-UTR)
- 3'-end
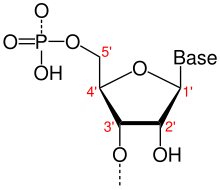
- One of two ends of a single linear strand of DNA or RNA, specifically the end at which the chain of nucleotides terminates at the third carbon atom in the furanose ring of deoxyribose or ribose (i.e. the terminus at which the 3' carbon is not attached to another nucleotide via a phosphodiester bond; in vivo, the 3' carbon is often still bonded to a hydroxyl group). By convention, sequences and structures positioned nearer to the 3'-end relative to others are referred to as downstream. Contrast 5'-end.
- 5' cap
- A specially altered nucleotide attached to the 5'-end of some primary RNA transcripts as part of the set of post-transcriptional modifications which convert raw transcripts into mature RNA products. The precise structure of the 5' cap varies widely by organism; in eukaryotes, the most basic cap consists of a methylated guanine nucleoside bonded to the triphosphate group that terminates the 5'-end of an RNA sequence. Among other functions, capping helps to regulate the export of mature RNAs from the nucleus, prevent their degradation by exonucleases, and promote translation in the cytoplasm. Mature mRNAs can also be decapped.
- 5' untranslated region (5'-UTR)
- 5-bromodeoxyuridine
- See bromodeoxyuridine.
- 5'-end
- One of two ends of a single linear strand of DNA or RNA, specifically the end at which the chain of nucleotides terminates at the fifth carbon atom in the furanose ring of deoxyribose or ribose (i.e. the terminus at which the 5' carbon is not attached to another nucleotide via a phosphodiester bond; in vivo, the 5' carbon is often still bonded to a phosphate group). By convention, sequences and structures positioned nearer to the 5'-end relative to others are referred to as upstream. Contrast 3'-end.
- 5-methyluracil
- See thymine.
A
[edit]- acentric
- (of a linear chromosome or chromosome fragment) Having no centromere.[2]
- acetyl coenzyme A (acetyl-CoA)
- A biochemical compound consisting of a coenzyme A molecule to which an acetyl group (–COCH
3) is attached via a high-energy thioester bond. Acetylation of coenzyme A occurs as part of the metabolism of proteins, carbohydrates (glycolysis), and fatty acids (beta oxidation), after which it participates as an energy carrier in several important biochemical pathways, notably the citric acid cycle, in which hydrolysis of the acetyl group releases energy which is ultimately captured in 11 ATP and one GTP. - acetylation
- The covalent attachment of an acetyl group (–COCH
3) to a chemical compound, protein, or other biomolecule via an esterification reaction with acetic acid, either spontaneously or by enzymatic catalysis. Acetylation plays important roles in several metabolic pathways and in histone modification. Contrast deacetylation. - acetyltransferase
- Any of a class of transferase enzymes which catalyze the covalent bonding of an acetyl group (–COCH
3) to another compound, protein, or biomolecule, a process known as acetylation. - acrocentric
- (of a linear chromosome or chromosome fragment) Having a centromere positioned very close to one end of the chromosome, as opposed to at the end or in the middle.[2]
- action potential
- The local change in voltage that occurs when the membrane potential of a specific location along the membrane of a cell rapidly depolarizes, such as when a nerve impulse is transmitted between neurons.
- activation
- See upregulation.
- activator
- A type of transcription factor that increases the transcription of a gene or set of genes. Most activators work by binding to a specific sequence located within or near an enhancer or promoter and facilitating the binding of RNA polymerase and other transcription machinery in the same region. See also coactivator; contrast repressor.
- active site
- The region of an enzyme to which one or more substrate molecules bind, causing the substrate or another molecule to undergo a chemical reaction. This region usually consists of one or more amino acid residues (commonly three or four) which, when the enzyme is folded properly, are able to form temporary chemical bonds with the atoms of the substrate molecule; it may also include one or more additional residues which, by interacting with the substrate, are able to catalyze a specific reaction involving the substrate. Though the active site constitutes only a small fraction of all the residues comprising the enzyme, its specificity for particular substrates and reactions is responsible for the enzyme's biological function.
- active transport
- Transport of a substance (such as a protein or drug) across a membrane against a concentration gradient. Unlike passive transport, active transport requires an expenditure of energy.
- adenine (A)
- A purine nucleobase used as one of the four standard nucleobases in both DNA and RNA molecules. Adenine forms a base pair with thymine in DNA and with uracil in RNA.
- adenosine (A)
- One of the four standard nucleosides used in RNA molecules, consisting of an adenine base with its N9 nitrogen bonded to the C1 carbon of a ribose sugar. Adenine bonded to deoxyribose is known as deoxyadenosine, which is the version used in DNA.
- adenosine diphosphate (ADP)
- adenosine monophosphate (AMP)
- adenosine triphosphate (ATP)
- An organic compound derived from adenine that functions as the major source of energy for chemical reactions inside living cells. It is found in all forms of life and is often referred to as the "molecular currency" of intracellular energy transfer.
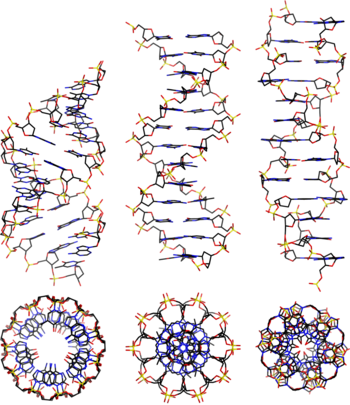
- A-DNA
- One of three main biologically active structural conformations of the DNA double helix, along with B-DNA and Z-DNA. The A-form helix has a right-handed twist with 11 base pairs per full turn, only slightly more compact than B-DNA, but its bases are sharply tilted with respect to the helical axis. It is often favored in dehydrated conditions and within sequences of consecutive purine nucleotides (e.g. GAAGGGGA); it is also the primary conformation adopted by double-stranded RNA and RNA-DNA hybrids.[3]
- affected relative pair
- Any pair of organisms which are related genetically and both affected by the same trait. For example, two cousins who both have blue eyes are an affected relative pair since they are both affected by the allele that codes for blue eyes.
- alkaline lysis
- A laboratory method used in molecular biology to extract and isolate extrachromosomal DNA such as the DNA of plasmids (as opposed to genomic or chromosomal DNA) from certain cell types, commonly cultured bacterial cells.
- allele
- One of multiple alternative versions of an individual gene, each of which is a viable DNA sequence occupying a given position, or locus, on a chromosome. For example, in humans, one allele of the eye-color gene produces blue eyes and another allele of the same gene produces brown eyes.
- allosome
- Any chromosome that differs from an ordinary autosome in size, form, or behavior and which is responsible for determining the sex of an organism. In humans, the X chromosome and the Y chromosome are sex chromosomes.
- alpha helix (α-helix)
- A common structural motif in the secondary structures of proteins consisting of a right-handed helix conformation resulting from hydrogen bonding between amino acid residues which are not immediately adjacent to each other.
- alternative splicing
- A regulated phenomenon of eukaryotic gene expression in which specific exons or parts of exons from the same primary transcript are variably included within or removed from the final, mature messenger RNA transcript. A class of post-transcriptional modification, alternative splicing allows a single gene to code for multiple protein isoforms and greatly increases the diversity of proteins that can be produced by an individual genome. See also RNA splicing.
- amber
- One of three stop codons used in the standard genetic code; in RNA, it is specified by the nucleotide triplet UAG. The other two stop codons are named ochre and opal.
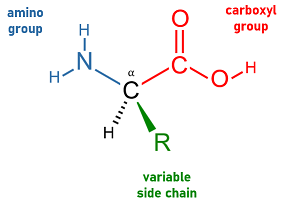
- amino acid
- Any of a class of organic compounds whose basic structural formula includes a central carbon atom bonded to amine and carboxyl functional groups and to a variable side chain. Out of nearly 500 known amino acids, a set of 20 are coded for by the standard genetic code and incorporated into long polymeric chains as the building blocks of peptides and hence of polypeptides and proteins. The specific sequences of amino acids in the polypeptide chains that form a protein are ultimately responsible for determining the protein's structure and function.
- amino terminus
- See N-terminus.
- aminoacyl-tRNA synthetase
- Any of a set of enzymes which catalyze the transesterification reaction that results in the attachment of a specific amino acid (or a precursor) to one of its cognate transfer RNA molecules, forming an aminoacyl-tRNA. Each of the 20 different amino acids used in the genetic code is recognized and attached by its own specific synthetase enzyme, and most synthetases are cognate to several different tRNAs according to their specific anticodons.
- aminoacyl-tRNA (aa-tRNA)
- A transfer RNA to which a cognate amino acid is chemically bonded; i.e. the product of a transesterification reaction catalyzed by an aminoacyl-tRNA synthetase. Aminoacyl-tRNAs bind to the aminoacyl site of the ribosome during translation.
- amplicon
- Any DNA or RNA sequence or fragment that is the source and/or product of an amplification reaction. The term is most frequently used to describe the numerous copied fragments that are the products of the polymerase chain reaction or ligase chain reaction, though it may also refer to sequences that are amplified naturally within a genome, e.g. by gene duplication.
- amplification
- The replication of a biomolecule, in particular the production of one or more copies of a nucleic acid sequence, known as an amplicon, either naturally (e.g. by spontaneous duplications) or artificially (e.g. by PCR), and especially implying many repeated replication events resulting in thousands, millions, or billions of copies of the target sequence, which is then said to be amplified.
- anaphase
- The stage of mitosis and meiosis that occurs after metaphase and before telophase, when the replicated chromosomes are segregated and each of the sister chromatids are moved to opposite sides of the cell.
- anaphase lag
- The failure of one or more pairs of sister chromatids or homologous chromosomes to properly migrate to opposite sides of the cell during anaphase of mitosis or meiosis due to a defective spindle apparatus. Consequently, both daughter cells are aneuploid: one is missing one or more chromosomes (creating a monosomy) while the other has one or more extra copies of the same chromosomes (creating a polysomy).
- aneucentric
- (of a linear chromosome or chromosome fragment) Having an abnormal number of centromeres, i.e. more than one.[4]
- aneuploidy
- The condition of a cell or organism having an abnormal number of one or more particular chromosomes (but excluding abnormal numbers of complete sets of chromosomes, which instead is known as euploidy).
- annealing
- The hybridization of two single-stranded nucleic acid molecules containing complementary sequences, creating a double-stranded molecule with paired nucleobases. The term is used in particular to describe steps in laboratory techniques such as polymerase chain reaction, where double-stranded DNA molecules are repeatedly denatured into single strands by heating and then exposed to cooler temperatures, causing the strands to reassociate with each other or with complementary primers. The exact temperature at which annealing occurs is strongly influenced by the length and specific sequence of the individual strands.
- antibody
- anticodon
- A series of three consecutive nucleotides within a transfer RNA which complement the three nucleotides of a codon within an mRNA transcript. During translation, each tRNA recruited to the ribosome contains a single anticodon triplet that pairs with its complementary codon from the mRNA sequence, allowing each codon to specify a particular amino acid to be added to the growing peptide chain. Anticodons containing inosine in the first position are capable of pairing with more than one codon due to a phenomenon known as wobble base pairing.
- antimetabolite
- Any molecule that functions as an antagonist to a metabolic process, limiting or inhibiting normal cellular metabolism; i.e. a metabolic poison.[4]
- antimitotic
- Any compound that suppresses normal mitosis in a cell or population of cells.[4]
- antioncogene
- A gene which helps to regulate cell growth and suppress tumors when functioning correctly, such that its absence or malfunction can result in uncontrolled cell growth and possibly cancer.[5] Compare oncogene.
- antiparallel
- The contrasting orientations of the two strands of a double-stranded nucleic acid (and more generally any pair of biopolymers) which are parallel to each other but with opposite directionality. For example, the two complementary strands of a DNA molecule run side-by-side but in opposite directions with respect to chemical numbering conventions, with one strand oriented 5'-to-3' and the other 3'-to-5'.
- antiporter
- A transport protein which works by exchanging two different ions or small molecules across a membrane in opposite directions, either at the same time or consecutively.[6]
- antisense
- See template strand.
- antisense RNA (asRNA)
- A single-stranded non-coding RNA molecule containing an antisense sequence that is complementary to a sense strand, such as a messenger RNA, with which it readily hybridizes, thereby inhibiting the sense strand's further activity (e.g. translation into protein). Many different classes of naturally occurring RNA such as siRNA function by this principle, making them potent gene silencers in various gene regulation mechanisms. Synthetic antisense RNA has also found widespread use in gene knockdown studies, and in practical applications such as antisense therapy.
- anucleate
- (of a cell or organism) Lacking a nucleus, i.e. a discrete, membrane-bound organelle enclosing the cell's genomic DNA, used especially of cells which normally have a nucleus but from which the nucleus has been removed (e.g. in artificial nuclear transfer), and also of specialized cell types that develop without nuclei despite that the cells of other tissues comprising the same organism ordinarily do have nuclei (e.g. mammalian erythrocytes).
- apical constriction
- The process by which contraction of the apical side of a cell (and often a corresponding expansion of the opposing basal side) causes the cell to assume a wedge-shaped morphology. The process is common during early development, where it is often coordinated across many adjacent cells of an epithelial layer simultaneously in order to generate bends or folds in developing tissues.
- apoptosis
- A highly regulated form of programmed cell death that occurs in multicellular organisms.
- aptamer
- Any artificial DNA, RNA, or XNA oligonucleotide molecule, single-stranded or double-stranded, which functions as a ligand by binding selectively to one or more specific target molecules, usually other nucleic acids or proteins, and often a family of such molecules. The term is used in particular to describe short nucleic acid fragments which have been randomly generated and then artificially selected in vitro by procedures such as SELEX. Aptamers are useful in the laboratory as antibody mimetics, particularly in applications where conventional protein antibodies are not appropriate.
- artificial gene synthesis
- A set of laboratory methods used in the de novo synthesis of a gene (or any other nucleic acid sequence) from free nucleotides, i.e. without relying on an existing template strand.
- assimilatory process
- Any process by which chemical compounds containing biologically relevant elements (e.g. carbon, hydrogen, oxygen, nitrogen, phosphorus, sulfur, selenium, iron, cobalt, nickel, copper, zinc, molybdenum, etc.) are uptaken by microorganisms and incorporated into complex biomolecules in order to synthesize various cellular components. In contrast, a dissimilatory process uses the energy released by decomposing exogenous molecules to power the cell's metabolism and excretes residual or toxic compounds out of the cell, instead of reusing them to build new molecules.
- aster
- In animal cells, a star-shaped system of non-kinetochore microtubules that radiates from a centrosome or from either of the poles of the mitotic spindle during the early stages of cell division.[6]
- asynapsis
- The failure of homologous chromosomes to properly pair with each other during meiosis.[4] Contrast synapsis and desynapsis.
- attached X
- A single monocentric chromosome containing two or more physically attached copies of the normal X chromosome as a result of either a natural internal duplication or any of a variety of genetic engineering methods. The resulting compound chromosome effectively carries two or more doses of all genes and sequences included on the X, yet functions in all other respects as a single chromosome, meaning that haploid 'XX' gametes (rather than the ordinary 'X' gametes) will be produced by meiosis and inherited by progeny. In mechanisms such as genic balance in which the sex of an organism is determined by the total dosage of X-linked genes, an abnormal 'XXY' zygote, fertilized by one XX gamete and one Y gamete, will develop into a female.
- autolysis
- The lysis or digestion of a cell by its own enzymes; or of a particular enzyme by another instance of the same enzyme. See also autophagy.
- autosome
- Any chromosome that is not an allosome and hence is not involved in the determination of the sex of an organism. Unlike the sex chromosomes, the autosomes in a diploid cell exist in pairs, with the members of each pair having the same structure, morphology, and genetic loci.
- autozygote
- A cell or organism that is homozygous for a locus at which the two homologous alleles are identical by descent, both having been derived from a single gene in a common ancestor.[4] Contrast allozygote.
- auxesis
- The growth of a multicellular organism due to an increase in the size of its cells rather than an increase in the number of cells.
- axenic
- Describing a cell culture in which only a single species, variety, or strain is present, and which is therefore entirely free of contaminating organisms including symbiotes and parasites.

B
[edit]- B chromosome
- Any supernumerary nuclear DNA molecule which is not a duplicate of nor homologous to any of the standard complement of normal "A" chromosomes comprising a genome. Typically very small and devoid of structural genes, B chromosomes are by definition not necessary for life. Though they occur naturally in many eukaryotic species, they are not stably inherited and thus vary widely in copy number even between closely related individuals.[4]
- back mutation
- A mutation that reverses the effect of a previous mutation which had inactivated a gene, thus restoring wild-type function.[7] See also reverse mutation.
- bacterial artificial chromosome (BAC)
- balancer chromosome
- base
- An abbreviation of nitrogenous base and nucleobase.
- base pair (bp)
- A pair of two nucleobases on complementary DNA or RNA strands which are loosely attracted to each other via hydrogen bonding, a type of non-covalent electrostatic interaction between individual atoms in the purine or pyrimidine rings of the complementing bases. This phenomenon, known as base pairing, is the mechanism underlying the hybridization that commonly occurs between nucleic acid polymers, allowing two single-stranded molecules to combine into a more energetically stable double-stranded molecule, as well as enabling certain individual strands to complement themselves. The ability of consecutive base pairs to stack one upon another contributes to the long-chain double helix structures observed in both double-stranded DNA and double-stranded RNA molecules.
- baseline
- A measure of the gene expression level of a gene or genes prior to a perturbation in an experiment, as in a negative control. Baseline expression may also refer to the expected or historical measure of expression for a gene.
- basic local alignment search tool (BLAST)
- A computer algorithm widely used in bioinformatics for aligning and comparing primary biological sequence information such as the nucleotide sequences of DNA or RNA or the amino acid sequences of proteins. BLAST programs enable scientists to quickly check for homology between two or more sequences by directly comparing the nucleotides or amino acids present at each position within each sequence; a common use is to search for matches between a specific query sequence and a digital sequence database such as a genome library, with the program returning a list of sequences from the database which resemble the query sequence above a specified threshold of similarity. Such comparisons can permit the identification of an organism from an unknown sample or the inference of evolutionary relationships between genes, proteins, or species.
- B-DNA
- The "standard" or classical structural conformation of the DNA double helix in vivo, thought to represent an average of the various distinct conformations assumed by very long DNA molecules under physiological conditions.[3] The B-form double helix has a right-handed twist with a diameter of 23.7 ångströms and a pitch of 35.7 ångströms or about 10.5 base pairs per full turn, such that each nucleotide pair is rotated 36° around the helical axis with respect to its neighboring pairs. See also A-DNA and Z-DNA.
- beta oxidation
- bidirectional replication
- A common mechanism of DNA replication in which two replication forks move in opposite directions away from the same origin; this results in a bubble-like region where the duplex molecule is locally separated into two single strands.[4]
- binary fission
- The separation of a single entity (e.g. a cell) into exactly two discrete entities closely resembling the original. The term refers in particular to a type of cell division used by prokaryotes such as bacteria, whereby a single parent cell divides evenly into two daughter cells which are genetically identical to each other and to the parent. Binary fission is preceded by replication of the parent cell's DNA, rapid growth of the cell wall, and various other processes which ensure even distribution of the cell's contents between the two progeny, but is generally a quicker and simpler process than the mitosis and cytokinesis that occur in eukaryotes.
- binding site
- bioassay
- Any analytical method that measures or qualifies the presence, effect, or potency of a substance within or upon a biological system, either directly or indirectly, e.g. by quantifying the concentration of a particular chemical compound within a sample obtained from living organisms, cells, or tissues, and ideally under controlled conditions that compare a sample subjected to an experimental treatment or manipulation with an unmanipulated sample, so as to permit inferences about the effect of the treatment upon some measured variable.[8]
- biochemical pathway
- bioenergetics
- biofilm
- A community of symbiotic microorganisms, especially bacteria, where cells produce and embed themselves within a slimy, sticky extracellular matrix composed of various high-molecular weight biopolymers, adhering to each other and sometimes also to a substratum, which may be a biotic or abiotic surface.[9] Many bacteria can exist either as independent single cells or switch to a physiologically distinct biofilm phenotype; those that create biofilms often do so in order to shelter themselves from harmful environments. Cells residing within biofilms can easily share nutrients and communicate, and subpopulations of cells may differentiate to perform specialized functions supporting the whole biofilm.[10]
- biomarker
- A measurable indicator of some biological state, especially a compound or biomolecule whose presence or absence in a biological system is a reliable sign of a normal or abnormal process, condition, or disease.[11] Things that may serve as biomarkers include direct measurements of the concentration of a particular compound or molecule in a tissue or fluid sample, or any other characteristic physiological, histological, or radiographic signal (e.g. a change in heart rate, or a distinct morphology under a microscope). They are regularly used as predictive or diagnostic tools in clinical medicine and laboratory research.
- biomolecular gradient
- Any difference in the concentration of biomolecules between two spaces within a biological system, whether intracellular, extracellular, across a membrane (e.g. between the cytoplasm on one side of the membrane and the external environment on the other), or between different cells or different parts of a tissue or organ system. Gradients of one kind or another drive virtually all biochemical processes occurring within and between cells, as natural systems tend to move toward a thermodynamic equilibrium where concentrations are uniformly distributed in all spaces and no gradients exist. Gradients thus cause chemical reactions to occur in particular directions, which can be used by cells to accomplish essential biological functions, including metabolic energy transfer, signal transduction, and the movement of specific ions and solutes into and out of cells and organelles. It is often necessary for cells to continuously regenerate gradients such as membrane potentials in order to permit these processes to continue.
- biomolecule
- Any molecule or chemical compound involved in or essential to one or more biological processes within a biological system, especially large macromolecules such as proteins, nucleic acids, lipids, and carbohydrates, but also broadly inclusive of smaller molecules such as vitamins, hormones, and biometals which are consumed or produced by biochemical reactions, often as part of biochemical pathways. Most biomolecules are organic compounds; some are produced naturally within cells or tissues (endogenous compounds), while others can only be obtained from the organism's environment (exogenous compounds).
- bivalent
- blast cell
- See precursor cell.
- blot
- Any of a variety of molecular biology methods by which electrophoretically or chromatographically separated DNA, RNA, or protein samples are transferred from a support medium such as a polyacrylamide or agarose gel onto an immobilizing carrier such as a nitrocellulose or PVDF membrane. Some methods involve the transfer of molecules by capillary action (e.g. Southern and northern blotting), while others rely on the transport of charged molecules by electrophoresis (e.g. western blotting). The transferred molecules are then visualized by colorant staining, by autoradiography, or by probing for specific sequences or epitopes with hybridization probes or antibodies bound to chemiluminescent reporters.[4]
- blunt end
- A term used to describe the end of a double-stranded DNA molecule where the terminal nucleobases on each strand are base-paired with each other, such that neither strand has a single-stranded "overhang" of unpaired bases, in contrast to a so-called "sticky end", where an overhang is created by one strand being one or more bases longer than the other. Blunt ends and sticky ends are relevant when ligating multiple DNA molecules, e.g. in restriction cloning, because sticky-ended molecules will not readily anneal to each other unless they have matching overhangs; blunt-ended molecules do not anneal in this way, so special procedures must be used to ensure that fragments with blunt ends are joined in the correct places.
- bromodeoxyuridine (BUDR, BrdU)
C
[edit]- cadastral gene
- A regulatory gene that restricts the expression of other genes to specific tissues or body parts in an organism, typically by producing gene products which variably inhibit or permit transcription of the other genes in different cell types.[4] The term is used most commonly in plant genetics.
- cadherin
- Any of a class of transmembrane proteins which are dependent on calcium ions (Ca2+) and whose extracellular domains function as mediators of cell–cell adhesion at adherens junctions in eukaryotic tissues.
- callus
- An unorganized mass of parenchymal cells that forms naturally at the site of wounds in plant tissues, and which is commonly artificially induced to form in plant tissue culture as a means of initiating somatic embryogenesis.[8]
- candidate gene
- A gene whose location on a chromosome is associated with a particular phenotype (often a disease-related phenotype), and which is therefore suspected of causing or contributing to the phenotype. Candidate genes are often selected for study based on a priori knowledge or speculation about their functional relevance to the trait or disease being researched.
- canonical sequence
- See consensus sequence.
- carbohydrate
- Any of a class of organic compounds having the generic chemical formula (CH
2O)
n, and one of several major classes of biomolecules found universally in biological systems. Carbohydrates include individual monosaccharides as well as larger polymeric oligosaccharides and polysaccharides, in which multiple monosaccharide monomers are joined by glycosidic bonds.[8] Abundant and ubiquitous, these compounds are involved in numerous essential biochemical processes and pathways; they are widely used as an energy source for cellular metabolism, as a form of energy storage, as signaling molecules, and as biomarkers to label or modify the activity of other molecules. Carbohydrates are often colloquially described as "sugars"; the prefix glyco- indicates a compound or process containing or involving carbohydrates, and the suffix -ose usually signifies that a compound is a carbohydrate or a derivative. - carboxyl terminus
- See C-terminus.
- carrier protein
- 1. A membrane protein that functions as a transporter, binding to a solute and facilitating its movement across the membrane by undergoing a series of conformational changes.[6]
- 2. A protein to which a specific ligand or hapten has been conjugated and which thereby carries an antigen capable of eliciting an antibody response.[12]
- 3. A protein which is included in an assay at high concentrations in order to prevent non-specific interactions of the assay's reagents with vessel surfaces, sample components, or other reagents.[12] For example, in many blotting techniques, albumin is intentionally allowed to bind non-specifically to the blotted membrane prior to fluorescent labelling, so as to "block" potential off-target binding of the fluorophore to the membrane, which might otherwise cause background fluorescence that obscures genuine signal from the target.
- caspase
- cassette
- A pre-existing nucleic acid sequence or construct, especially a DNA vector with an annotated sequence and precisely positioned regulatory elements, into which one or more fragments can be readily inserted or recombined by various genetic engineering methods. Recombinant plasmid vectors containing reliable promoters, origins of replication, and antibiotic resistance genes are commercially manufactured as cassettes to allow scientists to easily swap genes of interest into and out of an active "slot" or locus within the plasmid. See also multiple cloning site.
- CCAAT box
- A highly conserved regulatory DNA sequence located approximately 75 base pairs upstream (i.e. -75) of the transcription start site for many eukaryotic genes.[2]
- cDNA
- See complementary DNA.
- cell
- The basic structural and functional unit of which all living organisms are composed, essentially a self-replicating ball of protoplasm surrounded by a surface membrane which separates the interior from the external environment, thus providing a protected space in which the carefully controlled chemical reactions necessary to sustain biological processes can be carried out unperturbed. Unicellular organisms are composed of a single autonomous cell, whereas multicellular organisms consist of numerous cells cooperating together, with individual cells more or less specialized or differentiated to serve particular functions.[12] Cells vary widely in size, shape, and substructure, particularly between prokaryotes and eukaryotes. The typical cell is microscopic, averaging 1 to 20 micrometres (μm) in diameter, though they may range in size from 0.1 μm to more than 20 centimetres in diameter for the eggs laid by some birds and reptiles, which are highly specialized single-celled ova.[8]
- cell biology
- The branch of biology that studies the structures, functions, processes, and properties of biological cells, the self-contained units of life common to all living organisms.
- cell compartmentalization
- The subdivision of the interior of a cell into distinct, usually membrane-bound compartments, including the nucleus and organelles (endoplasmic reticulum, mitochondria, chloroplasts, intracellular vesicles, etc.), a defining feature of the Eukarya.[8]
- cell cortex
- A specialized layer of cytoplasmic proteins lining the inner face of the cell membrane in most eukaryotic cells, composed primarily of actin microfilaments and myosin motor proteins and usually 100–1000 nanometres thick, which functions as a modulator of membrane behavior and cell surface properties.
- cell counting
- The process of determining the number of cells within a biological sample or culture by any of a variety of methods. Counting cells is an important aspect of cytometry used widely in research and clinical medicine. It is generally achieved by using a manual or digital cytometer to count the number of cells present in small fractions of a sample, and then extrapolating to estimate the total number present in the entire sample. The resulting quantification is typically expressed as a density or concentration, i.e. the number of cells per unit area or volume.
- cell culture
- The process by which living cells are grown and maintained, or "cultured", under carefully controlled conditions, generally outside of their natural environment. Optimal growth conditions vary widely for different cell types but usually consist of a suitable vessel (e.g. a culture tube or Petri dish) containing a specifically formulated substrate or growth medium that supplies all of the nutrients essential for life (amino acids, carbohydrates, vitamins, minerals, etc.) plus any desirable growth factors and hormones, permits gas exchange (if necessary), and regulates the environment by maintaining consistent physico-chemical properties (pH, osmotic pressure, temperature, etc.). Some cell types require a solid surface to which they can adhere in order to reproduce, whereas others can be grown while floating freely in a liquid or gelatinous suspension. Most cells have a genetically determined reproduction limit, but immortalized cells will divide indefinitely if provided with optimal conditions.
- cell cycle
- cell division
- The separation of an individual parent cell into two daughter cells by any process. Cell division generally occurs by a complex, carefully structured sequence of events involving the reorganization of the parent cell's internal contents, the physical cleaving of the cytoplasm and plasma membrane, and the even distribution of contents between the two resulting cells, so that each ultimately contains approximately half of the original cell's starting material. It usually implies reproduction via the replication of the parent cell's genetic material prior to division, though cells may also divide without replicating their DNA. In prokaryotic cells, binary fission is the primary form of cell division. In eukaryotic cells, asexual division occurs by mitosis and cytokinesis, while specific lineages of cells reserved for sexual reproduction can additionally divide by meiosis.[6]
- cell fusion
- The merging or coalescence of two or more cells into a single cell, as occurs in the fusion of gametes to form a zygote. Generally this occurs by the destabilization of each cell's plasma membrane and the formation of cytoplasmic bridges between them which then expand until the two cytoplasms are completely mixed; intercellular structures or organelles such as nuclei may or may not fuse as well. Some cells can be artificially induced to fuse with each other by treating them with a fusogen such as polyethylene glycol or by passing an electric current through them.[8]
- cell membrane
- The selectively permeable membrane surrounding all prokaryotic and eukaryotic cells, defining the outermost boundary of the cell and physically separating the cytoplasm from the extracellular environment.[13] Like all membranes, the cell membrane is a flexible, fluid, sheet-like phospholipid bilayer with membrane proteins, carbohydrates, and numerous other molecules embedded within or interacting with it from both sides. Embedded molecules often have freedom to move laterally alongside the membrane's lipids. Though the cell membrane can be freely crossed by many ions, small organic molecules, and water, most other substances require active transport through special pores or channels or by endocytosis or exocytosis in order to enter or exit the cell, especially very large or electrically charged molecules such as proteins and nucleic acids. Besides regulating the transport of substances into and out of the cell, the cell membrane creates an organized interior space in which to perform life-sustaining activities and plays fundamental roles in all of the cell's interactions with its environment, making it important in cell signaling, motility, defense, and division, among numerous other processes.
- cell physiology
- The study of the various biological activities and biochemical processes which sustain life inside cells, particularly (but not necessarily limited to) those related to metabolism and energy transfer, growth and reproduction, and the ordinary processes of the cell cycle.
- cell polarity
- The spatial variation within a cell, i.e. the existence of differences in shape, structure, or function between different parts of the same cell. Almost all cell types exhibit some form of polarity, often along an invisible axis which defines opposing sides or poles where the variation is most extreme. Having internal polarity permits cells to accomplish specialized functions such as signal transduction or to serve as epithelial cells which must perform different tasks on different sides, or facilitates cell migration or division.
- cell signaling
- The diverse set of processes by which cells transmit information to and receive information from themselves, other cells, or their environment. Signal transduction occurs in all cell types, prokaryotic and eukaryotic, and is of critical importance to the cell's ability to navigate and survive its physical surroundings. Countless mechanisms of signaling have evolved in different organisms, which are often categorized according to the proximity between sender and recipient (autocrine, intracrine, juxtacrine, paracrine, or endocrine).
- cell surface receptor
- Any of a class of receptor proteins embedded within or attached to the external surface of the cell membrane, with one or more binding sites facing the extracellular environment and one or more effector sites that couple the binding of a particular ligand to an intracellular event or process. Cell surface receptors are a primary means by which environmental signals are received by the cell and transmitted across the membrane into the cell interior. Some may also bind exogenous ligands and transport them into the cell in a process known as receptor-mediated endocytosis.[14]
- cell wall
- A tough, variously flexible or rigid layer of polysaccharide or glycoprotein polymers surrounding some cell types immediately outside of the cell membrane, including plant cells and most prokaryotes, which functions as an additional protective and selective barrier and gives the cell a definite shape and structural support. The chemical composition of the cell wall varies widely between taxonomic groups, and even between different stages of the cell cycle: in land plants it consists primarily of cellulose, hemicellulose, and pectin, while algae make use of carrageenan and agar, fungi use chitin, and bacterial cell walls contain peptidoglycan.
- cell-free DNA (cfDNA)
- Any DNA molecule that exists outside of a cell or nucleus, freely floating in an extracellular fluid such as blood plasma.
- cellular
- Of, relating to, consisting of, produced by, or resembling a cell or cells.
- cellular differentiation
- See differentiation.
- cellular immunity
- A class of immune response that does not rely on the production of antibodies but rather the activation of specific cell types such as phagocytes or cytotoxic T-lymphocytes, or the secretion of various cytokines from cells, in response to an antigen.
- cellular noise
- Any apparently random variability observed in quantities measured in cell biology, particularly those pertaining to gene expression levels.[15]
- cellular reprogramming
- The conversion of a terminally differentiated cell from one tissue-specific cell type to another. This involves dedifferentiation to a pluripotent state; an example is the conversion of mouse somatic cells to an undifferentiated embryonic state, which relies on the transcription factors Oct4, Sox2, Myc, and Klf4.[16]
- cellular senescence
- centimorgan (cM)
- A unit for measuring genetic linkage defined as the distance between chromosomal loci for which the expected average number of intervening chromosomal crossovers in a single generation is 0.01. Though not an actual measure of physical distance, it is used to infer the actual distance between two loci based on the apparent likelihood of a crossover occurring between them in any given meiotic division.
- central dogma of molecular biology
- A generalized framework for understanding the flow of genetic information between macromolecules within biological systems. The central dogma outlines the fundamental principle that the sequence information encoded in the three major classes of biopolymer—DNA, RNA, and protein—can only be transferred between these three classes in certain ways, and not in others: specifically, information transfer between the nucleic acids and from nucleic acid to protein is possible, but transfer from protein to protein, or from protein back to either type of nucleic acid, is impossible and does not occur naturally.
- centriole
- A cylindrical organelle composed of microtubules, present only in certain eukaryotes. A pair of centrioles migrate to and define the two opposite poles of a dividing cell where, as part of a centrosome, they initiate the growth of the spindle apparatus.
- centromere
- A specialized DNA sequence within a chromosome that links a pair of sister chromatids. The primary function of the centromere is to act as the site of assembly for kinetochores, protein complexes which direct the attachment of spindle fibers to the centromere and facilitate segregation of the chromatids during mitosis or meiosis.
- centromeric index
- The proportion of the total length of a chromosome encompassed by its short arm, typically expressed as a percentage; e.g. a chromosome with a centromeric index of 15 is acrocentric, with a short arm comprising only 15% of its overall length.[4]
- centrosome
- cfDNA
- See cell-free DNA.
- channel protein
- A type of transmembrane protein whose shape forms an aqueous pore in a membrane, permitting the passage of specific solutes, often small ions, across the membrane in either or both directions.[6]
- Chargaff's rules
- A set of axioms which state that, in the DNA of any chromosome, species, or organism, the total number of adenine (A) residues will be approximately equal to the total number of thymine (T) residues, and the number of guanine (G) residues will be equal to the number of cytosine (C) residues; accordingly, the total number of purines (A + G) will equal the total number of pyrimidines (T + C). These observations illustrate the highly specific nature of the complementary base-pairing that occurs in all duplex DNA molecules: even though non-standard pairings are technically possible, they are exceptionally rare because the standard ones are strongly favored in most conditions. Still, the 1:1 equivalence is seldom exact, since at any given time nucleobase ratios are inevitably distorted to some small degree by unrepaired mismatches, missing bases, and non-canonical bases. The presence of single-stranded DNA polymers also alters the proportions, as an individual strand may contain any number of any of the bases.
- charged tRNA
- A transfer RNA to which an amino acid has been attached; i.e. an aminoacylated tRNA. Uncharged tRNAs lack amino acids.[4]
- chDNA
- See chloroplast DNA.
- chemiosmosis
- chemokinesis
- A non-directional, random change in the movement of a molecule, cell, or organism in response to a chemical stimulus, e.g. a change in speed resulting from exposure to a particular chemical compound.
- chemotaxis
- A directed, non-random change in the movement of a molecule, cell, or organism in response to a chemical stimulus, e.g. towards or away from an area with a high concentration of a particular chemical compound.[6]
- chiasma
- A cross-shaped junction that forms the physical point of contact between two non-sister chromatids belonging to homologous chromosomes during synapsis. As well as ensuring proper segregation of the chromosomes, these junctions are also the breakpoints at which chromosomal crossover may occur during mitosis or meiosis, which results in the reciprocal exchange of DNA between the synapsed chromatids.
- chimerism
- The presence of two or more populations of cells with distinct genotypes in an individual organism, known as a chimera, which has developed from the fusion of cells originating from separate zygotes; each population of cells retains its own genome, such that the organism as a whole is a mixture of genetically non-identical tissues. Genetic chimerism may be inherited (e.g. by the fusion of multiple embryos during pregnancy) or acquired after birth (e.g. by allogeneic transplantation of cells, tissues, or organs from a genetically non-identical donor); in plants, it can result from grafting or errors in cell division. It is similar to but distinct from mosaicism.
- chloroplast
- A type of small, lens-shaped plastid organelle found in the cells of green algae and plants which contains light-sensitive photosynthetic pigments and in which the series of biochemical reactions that comprise photosynthesis takes place. Like mitochondria, chloroplasts are bound by a double membrane, contain their own internal circular DNA molecules from which they direct transcription of a unique set of genes, and replicate independently of the nuclear genome.[8][12]
- chloroplast DNA (cpDNA, chDNA, ctDNA)
- The set of DNA molecules contained within chloroplasts, a type of photosynthetic plastid organelle located within the cells of some eukaryotes such as plants and algae, representing a semi-autonomous genome separate from that within the cell's nucleus. Like other types of plastid DNA, cpDNA usually exists in the form of small circular plasmids.
- chondriome
- The complete set of mitochondria or of mitochondrial DNA within a cell, tissue, organism, or species.
- chromatid
- One copy of a newly copied chromosome, which is joined to the original chromosome by a centromere. Paired copies of the same individual chromosome are known as sister chromatids.
- chromatin
- A complex of DNA, RNA, and protein found in eukaryotic cells that is the primary substance comprising chromosomes. Chromatin functions as a means of packaging very long DNA molecules into highly organized and densely compacted shapes, which prevents the strands from becoming tangled, reinforces the DNA during cell division, helps to prevent DNA damage, and plays an important role in regulating gene expression and DNA replication.
- chromatin immunoprecipitation (ChIP)
- chromocenter
- A central amorphous mass of polytene chromosomes found in the nuclei of cells of the salivary glands in Drosophila larvae and resulting from the fusion of heterochromatic regions surrounding the centromeres of the somatically paired chromosomes, with the distal euchromatic arms radiating outward.[4]
- chromomere
- A region of a chromosome that has been locally compacted or coiled into chromatin, conspicuous under a microscope as a "bead", node, or dark-staining band, especially when contrasted with nearby uncompacted strings of DNA.
- chromosomal crossover
- chromosomal DNA
- DNA contained in chromosomes, as opposed to extrachromosomal DNA. The term is generally used synonymously with genomic DNA.
- chromosomal duplication
- The duplication of an entire chromosome, as opposed to a segment of a chromosome or an individual gene.
- chromosomal instability
- chromosome
- A nuclear DNA molecule containing part or all of the genetic material of an organism. Chromosomes may be considered a sort of molecular "package" for carrying DNA within the nucleus of cells and, in most eukaryotes, are composed of long strands of DNA coiled with packaging proteins which bind to and condense the strands to prevent them from becoming an unmanageable tangle. Chromosomes are most easily distinguished and studied in their completely condensed forms, which only occur during cell division. Some simple organisms have only one chromosome made of circular DNA, while most eukaryotes have multiple chromosomes made of linear DNA.
- chromosome condensation
- The process by which eukaryotic chromosomes become shorter, thicker, denser, and more conspicuous under a microscope during prophase due to systemic coiling and supercoiling of chromatic strands of DNA in preparation for cell division.
- chromosome segregation
- The process by which sister chromatids or paired homologous chromosomes separate from each other and migrate to opposite sides of the dividing cell during mitosis or meiosis.
- chromosome walking
- See primer walking.
- cilium
- A slender, thread-like, membrane-bound projection extending from the surface of a eukaryotic cell, longer than a microvillus but shorter than a flagellum. Most eukaryotic cells have at least one primary cilium serving sensory or signaling functions; some cells employ thousands of motile cilia covering their entire surface in order to achieve locomotion or to move extracellular material past the cell.
- circular DNA
- Any DNA molecule, single-stranded or double-stranded, which forms a continuous closed loop without ends; e.g. bacterial chromosomes, mitochondrial and plastid DNA, as well as many other varieties of extrachromosomal DNA, including plasmids and some viral DNA. Contrast linear DNA.
- circulating tumor DNA (ctDNA)
- Any extracellular DNA fragments derived from tumor cells which are circulating freely in the bloodstream.
- cis
- On the same side; adjacent to; acting from the same molecule. Contrast trans.
- cis-acting
- Affecting a gene or sequence on the same nucleic acid molecule. A locus or sequence within a particular DNA molecule such as a chromosome is said to be cis-acting if it influences or acts upon other sequences located within short distances (i.e. physically nearby, usually but not necessarily downstream) on the same molecule or chromosome; or, in the broadest sense, if it influences or acts upon other sequences located anywhere (not necessarily within a short distance) on the same chromosome of a homologous pair. Cis-acting factors are often involved in the regulation of gene expression by acting to inhibit or to facilitate transcription. Contrast trans-acting.
- cis-dominant mutation
- A mutation occurring within a cis-regulatory element (such as an operator) which alters the functioning of a nearby gene or genes on the same chromosome. Cis-dominant mutations affect the expression of genes because they occur at sites that control transcription rather than within the genes themselves.
- cisgenesis
- cis-regulatory element (CRE)
- Any sequence or region of non-coding DNA which regulates the transcription of nearby genes (e.g. a promoter, operator, silencer, or enhancer), typically by serving as a binding site for one or more transcription factors. Contrast trans-regulatory.
- cisterna
- Any of a class of flattened, membrane-bound vesicles or saccules of the smooth and rough endoplasmic reticulum and the Golgi apparatus. By traveling through one or more cisternae, each of which contains a distinct set of enzymes, newly created proteins and polysaccharides undergo chemical modifications such as phosphorylation and glycosylation, which are used as packaging signals to direct their transport to specific destinations within the cell.[17]
- cistron
- citric acid cycle
- classical genetics
- The branch of genetics based solely on observation of the visible results of reproductive acts, as opposed to that made possible by the modern techniques and methodologies of molecular biology. Contrast molecular genetics.
- cleavage
- cleavage furrow
- A trough-like indentation in the surface of the parent cell, often conspicuous when viewed through a microscope, that initiates the cleavage of the cytoplasm (cytokinesis) as the contractile ring begins to narrow during cell division.
- clonal selection
- cloning
- The process of producing, either naturally or artificially, individual organisms or cells which are genetically identical to each other. Clones are the result of all forms of asexual reproduction, and cells that undergo mitosis produce daughter cells that are clones of the parent cell and of each other. Cloning may also refer to biotechnology methods which artificially create copies of organisms or cells, or, in molecular cloning, copies of DNA fragments or other molecules.
- closed chromatin
- See heterochromatin.
- coactivator
- A type of coregulator that increases the expression of one or more genes by binding to an activator.
- coding strand
- The strand of a double-stranded DNA molecule whose nucleotide sequence corresponds directly to that of the RNA transcript produced during transcription (except that thymine bases are substituted with uracil bases in the RNA molecule). Though it is not itself transcribed, the coding strand is by convention the strand used when displaying a DNA sequence because of the direct analogy between its sequence and the codons of the RNA product. Contrast template strand; see also sense.
- codon
- A series of three consecutive nucleotides in a coding region of a nucleic acid sequence. Each of these triplets codes for a particular amino acid or stop signal during protein synthesis. DNA and RNA molecules are each written in a language using four "letters" (four different nucleobases), but the language used to construct proteins includes 20 "letters" (20 different amino acids). Codons provide the key that allows these two languages to be translated into each other. In general, each codon corresponds to a single amino acid (or stop signal). The full set of codons is called the genetic code.
- codon usage bias
- The preferential use of a particular codon to code for a particular amino acid rather than alternative codons that are synonymous for the same amino acid, as evidenced by differences between organisms in the frequencies of the synonymous codons occurring in their coding DNA. Because the genetic code is degenerate, most amino acids can be specified by multiple codons. Nevertheless, certain codons tend to be overrepresented (and others underrepresented) in different species.
- coenocyte
- A multinucleate mass of cytoplasm bounded by a cell wall and resulting from continuous cytoplasmic growth and repeated nuclear division without cytokinesis, found in some species of algae and fungi, e.g. Vaucheria and Physarum.[8]
- coenzyme
- A relatively small, independent molecule which associates with a specific enzyme and participates in the reaction that the enzyme catalyzes, often by forming a covalent bond with the substrate. Examples include biotin, NAD+, and coenzyme A.[6]
- coenzyme A (CoA)
- cofactor
- Any non-protein organic compound capable of binding to or interacting with an enzyme. Cofactors are required for the initiation of catalysis.
- comparative genomic hybridization (CGH)
- competence
- complementarity
- A property of nucleic acid biopolymers whereby two polymeric chains or "strands" aligned antiparallel to each other will tend to form base pairs consisting of hydrogen bonds between the individual nucleobases comprising each chain, with each type of nucleobase pairing almost exclusively with one other type of nucleobase; e.g. in double-stranded DNA molecules, A pairs only with T and C pairs only with G. Strands that are paired in such a way, and the bases themselves, are said to be complementary. The degree of complementarity between two strands strongly influences the stability of the duplex molecule; certain sequences may also be internally complementary, which can result in a single strand binding to itself. Complementarity is fundamental to the mechanisms governing DNA replication, transcription, and DNA repair.
- complementary DNA (cDNA)
- DNA that is synthesized from a single-stranded RNA template (typically mRNA or miRNA) in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is produced both naturally by retroviruses and artificially in certain laboratory techniques, particularly molecular cloning. In bioinformatics, the term may also be used to refer to the sequence of an mRNA transcript expressed as its DNA coding strand counterpart (i.e. with thymine replacing uracil).
- compound X
- See attached X.
- conditional expression
- The controlled, inducible expression of a transgene, either in vitro or in vivo.
- confluence
- In cell culture, a measure of the proportion of the surface area of a culture vessel that is covered by adherent cells, commonly expressed as a percentage. A culture in which the entire surface is completely covered by a continuous monolayer, such that all cells are immediately adjacent to and in direct physical contact with other cells, with no gaps or voids, is said to be 100-percent confluent. Different cell lines exhibit differences in growth rate or gene expression depending on the degree of confluence. Because of contact inhibition, most show a significant reduction in the rate of cell division as they approach complete confluence, though some immortalized cells may continue to divide, expanding vertically rather than horizontally by stacking themselves on top of the parent cells, until all available nutrients are depleted.[8][12]
- conformation
- The three-dimensional spatial configuration of the atoms comprising a molecule or macromolecular structure.[8] The conformation of a protein is the physical shape into which its polypeptide chains arrange themselves during protein folding, which is not necessarily rigid and may change with the protein's particular chemical environment.
- conformational change
- A change in the spatial conformation or physical shape of a molecule or macromolecule such as a protein or nucleic acid, rarely spontaneously but more commonly as a result of some alteration in the molecule's chemical environment (e.g. temperature, pH, salt concentration, etc.) or an interaction with another molecule. Changes in the tertiary structures of proteins can affect whether or how strongly they bind ligands or substrates; inducing these changes is a common means (both naturally and artificially) of activating, inactivating, or otherwise controlling the function of many enzymes and receptor proteins.[12]
- congression
- The movement of chromosomes to the spindle equator during the prometaphase and metaphase stages of mitosis.[4]
- consensus sequence
- A calculated order of the most frequent residues (of either nucleotides or amino acids) found at each position in a common sequence alignment and obtained by comparing multiple closely related sequence alignments.
- conservative replication
- A hypothetical mode of DNA replication in which the two parental strands of the original double-stranded DNA molecule ultimately remain hybridized to each other at the end of the replication process, with the two daughter strands forming their own separate molecule; hence one molecule is composed of both of the starting strands while the other is composed of the two newly synthesized strands. This is in contrast to semiconservative replication, in which each molecule is a hybrid of one old and one new strand. See also dispersive replication.
- conserved sequence
- A nucleic acid or protein sequence that is highly similar or identical across many species or within a genome, indicating that it has remained relatively unchanged through a long period of evolutionary time.
- constitutive expression
- 1. The continuous transcription of a gene, as opposed to facultative expression, in which a gene is only transcribed as needed. A gene that is transcribed continuously is called a constitutive gene.
- 2. A gene whose expression depends only on the efficiency of its promoter in binding RNA polymerase,[4] and not on any transcription factors or other regulatory elements which might promote or inhibit its transcription.
- contact inhibition
- In cell culture, the phenomenon by which most normal eukaryotic cells adhering to a planar substratum cease to grow and divide upon reaching a critical cell density, usually as they approach full confluence or come into physical contact with other cells. As a result, many types of cells cultured on plates or in Petri dishes will continue to proliferate until they cover the whole surface of the culture vessel, at which point the rate of cell division abruptly decreases or is arrested entirely, thus forming a confluent monolayer with minimal overlap between neighboring cells, even if the nutrient medium remains plentiful, rather than stacking themselves on top of each other.[14] Transformed or neoplastic cells tend not to respond to cell density in the same way and may continue to proliferate at high densities.[8] This type of density-dependent inhibition of growth is similar to and may occur simultaneously with, but is nonetheless distinct from, the related phenomenon of contact inhibition of movement,[12] whereby moving cells respond to physical contact by temporarily stopping and then reversing their direction of locomotion away from the point of contact.
- contig
- A continuous sequence of genomic DNA generated by assembling cloned fragments by means of their overlapping sequences.[7]
- cooperativity
- A phenomenon observed in some enzymes, receptor proteins, and protein complexes which have multiple binding sites, whereby the binding of a ligand to one or more sites apparently increases or decreases the affinity of one or more other binding sites for other ligands. This concept highlights the sensitive nature of the chemistry that governs interactions between biomolecules: the strength and specificity of interactions between protein and ligand are influenced, sometimes substantially, by nearby interactions (often conformational changes) and by the local chemical environment in general. Cooperativity is frequently invoked to account for the non-linearity of data resulting from attempts to measure the association/dissociation constants of particular protein–protein interactions.[12]
- copy DNA (cDNA)
- See complementary DNA.
- copy error
- A mutation resulting from a mistake made during DNA replication.[4]
- copy-number variation (CNV)
- A phenomenon in which sections of a genome are repeated and the number of repeats varies between individuals in the population, usually as a result of duplication or deletion events that affect entire genes or sections of chromosomes. Copy-number variations play an important role in generating genetic variation within a population.
- coregulator
- A protein that works together with one or more transcription factors to regulate gene expression.
- corepressor
- A type of coregulator that reduces (represses) the expression of one or more genes by binding to and activating a repressor.
- cosmid
- cpDNA
- See chloroplast DNA.
- CpG island
- A region of a genome in which CpG sites occur repetitively or with high frequency.
- CpG site
- A sequence of DNA in which a cytosine nucleotide is immediately followed by a guanine nucleotide on the same strand in the 5'-to-3' direction; the "p" in CpG refers simply to the intervening phosphate group linking the two consecutive nucleotides.
- CRISPR gene editing
- crista
- Any of numerous folds or invaginations in the inner mitochondrial membrane, which give this membrane its characteristic wrinkled shape and increase the surface area across which aerobic gas exchange and supporting electron transport reactions can occur. Cristae are studded with proteins such as ATP synthase and various cytochromes.
- crossing over
- See chromosomal crossover.
- crosslink
- Any chemical bond or series of bonds, normal or abnormal, natural or artificial, that connects two or more polymeric molecules to each other, creating an even larger, often structurally rigid and mechanically durable macromolecular complex. Crosslinks may consist of covalent, ionic, or intermolecular interactions, or even extensive physical entanglements of molecules, and may be reversible or irreversible; in polymer chemistry the term is often used to describe macrostructures that form predictably in the presence of a specific reagent or catalyst. In molecular biology the usage generally implies abnormal bonding (whether naturally occurring or experimentally induced) between different biomolecules (or different parts of the same biomolecule) which are ordinarily separate, especially nucleic acids and proteins. Crosslinking of DNA may occur between nucleobases on opposite strands of a double-stranded DNA molecule (interstrand), or between bases on the same strand (intrastrand), specifically via the formation of covalent bonds that are stronger than the hydrogen bonds of normal base pairing; these are common targets of DNA repair pathways. Proteins are also susceptible to becoming crosslinked to DNA or to other proteins through bonds to specific surface residues, a process which is artificially induced in many laboratory methods such as fixation and which can be useful for studying interactions between proteins in their native states. Crosslinks are generated by a variety of exogenous and endogenous agents, including chemical compounds and high-energy radiation, and tend to interfere with normal cellular processes such as DNA replication and transcription, meaning their persistence usually compromises cell health.
- ctDNA
- 1. An abbreviation of circulating tumor DNA.
- 2. An abbreviation of chloroplast DNA.
- C-terminus
- The end of a linear chain of amino acids (i.e. a peptide) that is terminated by the free carboxyl group (–COOH) of the last amino acid to be added to the chain during translation. This amino acid is said to be C-terminal. By convention, sequences, domains, active sites, or any other structure positioned nearer to the C-terminus of the polypeptide or the folded protein it forms relative to others are described as downstream. Contrast N-terminus.
- cut
- C-value
- The total amount of DNA contained within a haploid nucleus (e.g. a gamete) of a particular organism or species, expressed in number of base pairs or in units of mass (typically picograms); or, equivalently, one-half the amount in a diploid somatic cell. For simple diploid eukaryotes the term is often used interchangeably with genome size, but in certain cases, e.g. in hybrid polyploids descended from parents of different species, the C-value may actually represent two or more distinct genomes contained within the same nucleus. C-values apply only to genomic DNA, and notably exclude extranuclear DNA.
- C-value enigma
- A term used to describe a diverse variety of questions regarding the immense variation in nuclear C-value or genome size among eukaryotic species, in particular the observation that genome size does not correlate with the perceived complexity of organisms, nor necessarily with the number of genes they possess; for example, many single-celled protists have genomes containing thousands of times more DNA than the human genome. This was considered paradoxical until the discovery that eukaryotic genomes consist mostly of non-coding DNA, which lacks genes by definition. The focus of the enigma has since shifted to understanding why and how eukaryotic genomes came to be filled with so much non-coding DNA, and why some genomes have a higher gene content than others.
- cyclic adenosine monophosphate (cAMP)
- cyclosis
- See cytoplasmic streaming.
- cytidine (C, Cyd)
- One of the four standard nucleosides used in RNA molecules, consisting of a cytosine base with its N9 nitrogen bonded to the C1 carbon of a ribose sugar. Cytosine bonded to deoxyribose is known as deoxycytidine, which is the version used in DNA.
- cytochemistry
- The branch of cell biology involving the detection and identification of various cellular structures and components, in particular their localization within cells, using techniques of biochemical analysis and visualization such as chemical staining and immunostaining, spectrophotometry and spectroscopy, radioautography, and electron microscopy.
- cytogenetics
- The branch of genetics that studies how chromosomes influence and relate to cell behavior and function, particularly during mitosis and meiosis.
- cytokine
- Any of a broad and loosely defined class of small proteins and peptides which have functions in intercellular signaling (primarily autocrine, paracrine, and endocrine pathways), typically by interacting with specific receptors on the exterior surface of cells.[6]
- cytokinesis
- The final stage of cell division in both mitosis and meiosis, usually immediately following the division of the nucleus, during which the cytoplasm of the parent cell is cleaved and divided approximately evenly between two daughter cells. In animal cells, this process occurs by the closing of a microfilament contractile ring in the equatorial region of the dividing cell. Contrast karyokinesis.
- cytology
- The study of the morphology, processes, and life history of living cells, particularly by means of light and electron microscopy.[8] The term is also sometimes used as a synonym for the broader field of cell biology.
- cytolysis
- See lysis.
- cytometer
- cytomics
- The interdisciplinary field that studies cell biology, cytology, and biochemistry at the level of an individual cell by making use of single-cell molecular techniques and advanced microscopy to visualize the interactions of cellular components in vivo.[18]
- cytoplasm
- cytoplasmic streaming
- The flow of the cytoplasm inside a cell, driven by forces exerted upon cytoplasmic fluids by the cytoskeleton. This flow functions partly to speed up the transport of molecules and organelles suspended in the cytoplasm to different parts of the cell, which would otherwise have to rely on passive diffusion for movement. It is most commonly observed in very large eukaryotic cells, for which there is a greater need for transport efficiency.
- cytoplast
- An enucleated eukaryotic cell; or all other cellular components besides the nucleus (i.e. the cell membrane, cytoplasm, organelles, etc.) considered collectively. The term is most often used in the context of nuclear transfer experiments, during which the cytoplast can sometimes remain viable in the absence of a nucleus for up to 48 hours.[14]
- cytosine (C)
- A pyrimidine nucleobase used as one of the four standard nucleobases in both DNA and RNA molecules. Cytosine forms a base pair with guanine.
- cytosol
- The soluble aqueous phase of the cytoplasm, in which small particles such as ribosomes, proteins, nucleic acids, and many other molecules are suspended or dissolved, excluding larger structures and organelles such as mitochondria, chloroplasts, lysosomes, and the endoplasmic reticulum.[8]


D
[edit]- daughter cell
- A cell resulting from the division of an initial progenitor, known as the parent cell. Generally two daughter cells are produced per division.[8]
- Denoising Algorithm based on Relevance network Topology
- An unsupervised algorithm that estimates an activity score for a pathway in a gene expression matrix, following a denoising step.[19]
- de novo mutation
- A spontaneous mutation in the genome of an individual organism that is new to that organism's lineage, having first appeared in a germ cell of one of the organism's parents or in the fertilized egg that develops into the organism; i.e. a mutation that was not present in either parent's genome.[4]
- de novo synthesis
- The assembly of a synthetic nucleic acid sequence from free nucleotides without relying on an existing template strand, i.e. de novo, by any of a variety of laboratory methods. De novo synthesis makes it theoretically possible to construct completely artificial molecules with no naturally occurring equivalent, and no restrictions on size or sequence. It is performed routinely in the commercial production of customized, made-to-order oligonucleotide sequences such as primers.
- deacetylation
- The removal of an acetyl group (–COCH
3) from a chemical compound, protein, or other biomolecule via hydrolysis of the covalent ester bond adhering it, either spontaneously or by enzymatic catalysis. Deacetylation is the opposite of acetylation. - decellularization
- degeneracy
- The redundancy of the genetic code, exhibited as the multiplicity of different codons that specify the same amino acid. For example, in the standard genetic code, the amino acid serine is specified by six unique codons (UCA, UCG, UCC, UCU, AGU, and AGC). Codon degeneracy accounts for the existence of synonymous mutations.
- degranulation
- The release of the contents of a secretory granule (usually antimicrobial or cytotoxic molecules) into an extracellular space by the exocytotic fusion of the granule with the cell's plasma membrane.[12]
- deletion
- A type of mutation in which one or more nucleotides are removed from a nucleic acid sequence.
- demethylation
- The removal of a methyl group (–CH
3) from a chemical compound, protein, or other biomolecule, either spontaneously or by enzymatic catalysis. Demethylation is the opposite of methylation; both reactions play important roles in numerous biochemical processes, including in regulating gene expression, as the methylation state of particular residues within particular proteins or nucleic acids can affect their structural conformation in a way that alters their affinity for other molecules, making transcription at nearby genetic loci more or less likely. - denaturation
- The process by which nucleic acids or proteins lose their quaternary, tertiary, and/or secondary structures, either reversibly or irreversibly, through the application of some external chemical or mechanical stress, e.g. by heating, agitation, or exposure to a strong acid or base, all of which can disrupt intermolecular forces such as hydrogen bonding and thereby change or destroy chemical activity. Denatured proteins may be both a cause and a consequence of cell death. Denaturation may also be a normal process; the denaturation of double-stranded DNA molecules, for example, which breaks the hydrogen bonds between base pairs and causes the separation of the duplex molecule into two single strands, is a necessary step in DNA replication and transcription and hence is routinely performed by enzymes such as helicases. The same mechanism is also fundamental to laboratory methods such as PCR.
- deoxyadenosine
- One of the four standard deoxyribonucleosides used in DNA molecules, consisting of an adenine base with its N9 nitrogen bonded to the C1 carbon of a deoxyribose sugar. Adenine bonded to ribose forms an alternate compound known simply as adenosine, which is used in RNA.
- deoxycytidine
- One of the four standard deoxyribonucleosides used in DNA molecules, consisting of a cytosine base with its N9 nitrogen bonded to the C1 carbon of a deoxyribose sugar. Cytosine bonded to ribose forms an alternate compound known simply as cytidine, which is used in RNA.
- deoxyguanosine
- One of the four standard deoxyribonucleosides used in DNA molecules, consisting of a guanine base with its N9 nitrogen bonded to the C1 carbon of a deoxyribose sugar. Guanine bonded to ribose forms an alternate compound known simply as guanosine, which is used in RNA.
- deoxyribonuclease (DNase)
- Any of a class of nuclease enzymes which catalyze the hydrolytic cleavage of phosphodiester bonds in DNA molecules, thereby severing strands of deoxyribonucleotides and causing the degradation of DNA polymers into smaller components. Compare ribonuclease.
- deoxyribonucleic acid (DNA)
- A polymeric nucleic acid molecule composed of a series of covalently linked deoxyribonucleotides, each of which incorporates one of four nucleobases: adenine (A), guanine (G), cytosine (C), and thymine (T). DNA is most often found in double-stranded form, which consists of two complementary, antiparallel nucleotide chains in which each of the nucleobases on each individual strand is paired via hydrogen bonding with one on the opposite strand; this structure commonly assumes the shape of a double helix. DNA can also exist in single-stranded form. By storing and encoding genetic information in the sequence of these nucleobases, DNA serves as the universal molecular basis of biological inheritance and the fundamental template from which all proteins, cells, and living organisms are constructed.
- deoxyribonucleotide
- A nucleotide containing deoxyribose as its pentose sugar component, and the monomer or subunit used to build deoxyribonucleic acid (DNA) molecules. Deoxyribonucleotides canonically incorporate any of four nitrogenous bases: adenine (A), guanine (G), cytosine (C), and thymine (T). Compare ribonucleotide.
- deoxyribose
- A monosaccharide pentose sugar derived from ribose by the replacement of the hydroxyl group attached to the C2 carbon with a single hydrogen atom. D-deoxyribose, in its cyclic ring form, is one of three main functional groups of deoxyribonucleotides and hence of deoxyribonucleic acid (DNA) molecules.
- deoxythymidine
- See thymidine.
- dephosphorylation
- The removal of a phosphate group, PO3−
4, from a chemical compound, protein, or other biomolecule, either spontaneously or by enzymatic catalysis. Dephosphorylation is the opposite of phosphorylation; both reactions are common molecular modifications involved in numerous biochemical pathways and processes, including in metabolism, where high-energy bonds to phosphate groups are used to transfer energy between molecules, and in the post-translational modification of proteins, where the phosphorylation state of particular residues can affect the protein's affinity for other molecules or function as a molecular signal. - depurination
- The spontaneous loss of one or more purine nucleobases (either adenine or guanine) from a nucleotide or nucleic acid molecule, either DNA or RNA, via the hydrolytic cleavage of the glycosidic bond linking base and sugar, releasing a free purine nucleobase and a nucleoside. Deoxyribonucleotides are especially prone to depurination. Loss of pyrimidine bases can also occur spontaneously but is far less common.
- derivatization
- The artificial modification of a molecule or protein with the intent of altering its solubility or other chemical properties so as to enable analysis (e.g. by mass spectroscopy or chromatography), or of labelling it by attaching a detectable chemical moiety (e.g. a fluorescent tag) to make it easier to identify and track in vivo. Molecules modified in this way are described as derivatives of their naturally occurring counterparts and are said to have been derivatized.[12]
- desmosome
- A specialized cell junction between neighboring epithelial cells in which the cells are held together by a network of keratin filaments and structural proteins bridging the gap between the plasma membranes.[12]
- destination vector
- desynapsis
- The failure of homologous chromosomes that have synapsed normally during pachynema to remain paired during diplonema. Desynapsis is usually caused by the improper formation of chiasmata.[4] Contrast asynapsis.
- developmental biology
- The branch of biology that studies the various processes and phenomena by which organisms (particularly multicellular eukaryotes but not necessarily excluding prokaryotes) grow and develop into mature forms capable of reproduction. In the broadest sense the field may encompass topics such as sexual and asexual reproduction, gametogenesis and sporogenesis, fertilization, embryogenesis, the renewal and differentiation of stem cells into specialized cell types, birth or hatching, metamorphosis, and the regeneration of mature tissues.
- diakinesis
- In meiosis, the fifth and final substage of prophase I, following diplonema and preceding metaphase I. During diakinesis, the chromosomes are further condensed, the two centrosomes reach opposite poles of the cell, and the spindle apparatus begins to extend from the poles to the equator.[4]
- dicentric
- (of a linear chromosome or chromosome fragment) Having two centromeres instead of the normal one.[2]
- differentiation
- The process by which a eukaryotic cell changes from one cell type to another, in particular from a non-specialized stem cell to a more specialized cell type which is then said to be differentiated. This usually occurs by a carefully regulated series of epigenetic modifications which change the specific set of genes expressed by the cell, turning certain genes off and others on, and (with few exceptions) almost never involves mutations to the nucleotide sequences themselves. These alterations to gene expression result in a cascade of phenotypic changes which can dramatically change the cell's size, shape, metabolism, membrane characteristics, and rate of division, and therefore its functions, behaviors, and responsiveness to signals, permitting multicellular organisms to create a huge variety of functionally distinct cell types from a single genome. Differentiation occurs repeatedly during an organism's development from a single-celled zygote into a complex multicellular system of tissues and cell types, and continues to some extent after the organism reaches maturity in order to repair and replace damaged and dying cells. In most cases differentiation is irreversible, though some cells may also undergo dedifferentiation in specific circumstances.
- dimer
- A molecular aggregate consisting of two subunits. The term is often used to describe a protein complex composed of two proteins, either the same protein (a homodimer) or different proteins (a heterodimer); or to an individual protein composed of two polypeptides. Compare monomer, trimer, and tetramer.
- dinucleotide
- A molecular dimer consisting of exactly two covalently linked nucleotides; or any two nucleotides which are immediately adjacent to each other on the same strand of a longer nucleic acid polymer.
- diploid
- (of a cell or organism) Having two homologous copies of each chromosome. Contrast haploid and polyploid.
- diplonema
- In meiosis, the fourth of the five substages of prophase I, following pachynema and preceding diakinesis. During diplonema, the synaptonemal complex disassembles and the paired homologous chromosomes begin to separate from one another, though they remain tightly bound at the chiasmata where crossover has occurred.
- direct repeat
- Any two or more repetitions of a specific sequence of nucleotides occurring in the same orientation (i.e. in precisely the same order and not inverted) and on the same strand, either separated by intervening nucleotides or not. An example is the sequence TACCGnnnnnnTACCG, in which TACCG occurs twice, though separated by six nucleotides that are not part of the repeated sequence. A direct repeat in which the repeats are immediately adjacent to each other is known as a tandem repeat.
- directionality
- The end-to-end chemical orientation of a single linear strand or sequence of a nucleic acid polymer or a polypeptide. The nomenclature used to indicate nucleic acid directionality is based on the chemical convention of identifying individual carbon atoms in the ribose or deoxyribose sugars of nucleotides, specifically the 5' carbon and 3' carbon of the pentose ring. The sequence of nucleotides in a polymeric chain may be read or interpreted in the 5'-to-3' direction – i.e. starting from the terminal nucleotide in which the 5' carbon is not connected to another nucleotide, and proceeding to the other terminal nucleotide, in which the 3' carbon is not connected to another nucleotide – or in the opposite 3'-to-5' direction. Most types of nucleic acid synthesis, including DNA replication and transcription, work exclusively in the 5'-to-3' direction, because the polymerases involved can only catalyze the addition of free nucleotides to the open 3'-end of the previous nucleotide in the chain. Because of this, the convention when writing any nucleic acid sequence is to present it in the 5'-to-3' direction from left to right. In double-stranded nucleic acids, the two paired strands must be oriented in opposite directions in order to base-pair with each other. Polypeptide directionality is similarly based on labeling the functional groups comprising amino acids, specifically the amino group, which forms the N-terminus, and the carboxyl group, which forms the C-terminus; amino acid sequences are assembled in the N-to-C direction during translation, and by convention are written in the same direction.
- disaccharide
- A carbohydrate composed of two monosaccharides (either the same or different) joined by a covalent glycosidic bond.[12] See also oligosaccharide and polysaccharide.
- dispersive replication
- dissimilatory process
- Any exergonic process of microbial catabolism by which redox-active chemical species participate in oxidation-reduction reactions (exchange of electrons) to provide the cell with energy needed for sustaining metabolic activities. External substances are absorbed by the cell from its environment and then decomposed to release energy, with the byproducts subsequently excreted out of the cell. This is in contrast to an assimilatory process, in which the atoms of the external substances are reused in the synthesis of biomolecules or the fabrication of cellular components.
- distance measure
- Any quantity used to measure the dissimilarity between the gene expression levels of different genes.[20]
- DNA
- See deoxyribonucleic acid.
- DNA barcoding
- A method of taxonomic identification in which short DNA sequences from one or more specific genes are isolated from unidentified samples and then aligned with sequences from a reference library in order to uniquely identify the species or other taxon from which the samples originated. The sequences used in the comparison are chosen carefully from genes that are both widely conserved and that show greater variation between species than within species, e.g. the cytochrome c oxidase gene for eukaryotes or certain ribosomal RNA genes for prokaryotes. These genes are present in nearly all living organisms but tend to evolve different mutations in different species, such that a unique sequence variant can be linked to one particular species, effectively creating a unique identifier akin to a retail barcode. DNA barcoding allows unknown specimens to be identified from otherwise indistinct tissues or body parts, where identification by morphology would be difficult or impossible, and the library of organismal barcodes is now comprehensive enough that even organisms previously unknown to science can often be phylogenetically classified with confidence. The simultaneous identification of multiple different species from a mixed sample is known as metabarcoding.
- DNA condensation
- The process of compacting very long DNA molecules into densely packed, orderly configurations such as chromosomes, either in vivo or in vitro.
- DNA fingerprinting
- DNA methylation
- DNA microarray
- A high-throughput technology used to measure expression levels of mRNA transcripts or to detect certain changes in nucleotide sequence. It consists of an array of thousands of microscopic spots of DNA oligonucleotides, called features, each containing picomoles of a specific DNA sequence. This can be a short section of a gene or any other DNA element, and is used as a probe to hybridize a cDNA, cRNA or genomic DNA sample (called a target) under high-stringency conditions. Probe-target hybridization is usually detected and quantified by fluorescence-based detection of fluorophore-labeled targets.
- DNA polymerase
- Any of a class of enzymes which synthesize DNA molecules from individual deoxyribonucleotides. DNA polymerases are essential for DNA replication and usually work in pairs to create identical copies of the two strands of an original double-stranded molecule. They build long chains of DNA by adding nucleotides one at a time to the 3'-end of a DNA strand, usually relying on the template provided by the complementary strand to copy the nucleotide sequence faithfully.
- DNA repair
- The set of processes by which a cell identifies and corrects structural damage or mutations in the DNA molecules that encode its genome. The ability of a cell to repair its DNA is vital to the integrity of the genome and the normal functionality of the organism.
- DNA replication
- The process by which a DNA molecule copies itself, producing two identical copies of one original DNA molecule.
- DNA sequencing
- The process of determining, by any of a variety of different methods and technologies, the order of the bases in the long chain of nucleotides that constitutes a sequence of DNA.
- DNA turnover
- Any mechanism by which DNA sequences are exchanged non-reciprocally (e.g. via gene conversion, transposition, or unequal crossing-over) that causes continual fluctuations in the copy number of DNA motifs during an organism's lifetime. Such mechanisms are often major drivers of speciation between populations.[14]
- DNA-binding domain (DBD)
- A protein domain containing at least one structural motif capable of recognizing and interacting with the nucleotides of a double-stranded or single-stranded DNA molecule. DNA-binding domains may bind to specific sequences or have a non-specific affinity for DNA. They are the primary functional components of DNA-binding proteins, including many transcription factors and regulatory proteins.
- DNA-binding protein (DBP)
- Any polypeptide or protein containing one or more domains capable of interacting chemically with one or more parts of a DNA molecule, and consequently having a specific or general affinity for single- and/or double-stranded DNA. DNA-binding activity often depends on the presence and physical accessibility of a specific nucleobase sequence, and mostly occurs at the major groove, since it exposes more of the functional groups which uniquely identify the bases. Binding is also influenced by the spatial conformation of the DNA chain and the occupancy of other proteins near the binding site; many proteins cannot bind to DNA without first undergoing conformational changes induced by interactions with other molecules.
- DNase
- See deoxyribonuclease.
- domain
- A discrete, usually continuous region of a protein, or the corresponding amino acid sequence of a polypeptide, which serves a particular function or is defined by particular physico-chemical properties (e.g. hydrophobic, polar, non-polar, globular, etc.),[12] and especially one which assumes a unique, recognizable spatial conformation as part of the protein's tertiary structure and which contributes to or defines its biological activity. Large proteins are generally composed of several domains linked together by short, intervening polypeptide sequences.[6] Domains are commonly grouped into classes with similar properties or functions, e.g. DNA-binding domains. More broadly, the term may also be used to refer to a discrete structural entity within any biomolecule, including functionally or compositionally distinct subregions of nucleic acid sequences and chromosomes.[14]
- donor vector
- dosage compensation
- Any mechanism by which organisms neutralize the large difference in gene dosage caused by the presence of differing numbers of sex chromosomes in the different sexes, thereby equalizing the expression of sex-linked genes so that the members of each sex receive the same or similar amounts of the products of such genes. An example is X-inactivation in female mammals.
- double helix
- The shape most commonly assumed by double-stranded nucleic acid molecules, resembling a ladder that has been twisted upon its long axis, with the rungs of the ladder consisting of paired nucleobases. This secondary structure is the most energetically stable conformation of the double-stranded forms of both DNA and RNA under most naturally occurring conditions, arising as a consequence of the primary structure of the phosphodiester backbone and the stacking of the nucleotides bonded to it. In B-DNA, the most common DNA variant found in nature, the double helix has a right-handed twist with about 10 base pairs per full turn, and the molecular geometry results in an alternating pattern of "grooves" of differing widths (a major groove and a minor groove) between the parallel backbones.
- double-strand break (DSB)
- The loss of continuity of the phosphate-sugar backbone in both strands of a double-stranded DNA molecule, in particular when the two breaks occur at sites that are directly across from or very close to each other on the complementary strands.[14] Contrast single-strand break.
- double-stranded
- Composed of two antiparallel, complementary nucleic acid molecules or strands (either DNA–DNA, RNA–RNA, or a DNA–RNA hybrid) which are held together by hydrogen bonds between the complementary nucleobases of each strand, known as base pairing. Compare single-stranded.
- double-stranded DNA (dsDNA)
- Any DNA molecule that is composed of two antiparallel, complementary deoxyribonucleotide polymers, known as strands, which are bonded together by hydrogen bonds between the complementary nucleobases. Though it is possible for DNA to exist as a single strand, it is generally more stable and more common in double-stranded form. In most cases, the complementary base pairing causes the twin strands to coil around each other in the shape of a double helix.
- double-stranded RNA (dsRNA)
- Any RNA molecule that is composed of two antiparallel, complementary ribonucleotide polymers, known as strands, which are bonded together by hydrogen bonds between the complementary nucleobases. Though RNA usually occurs in single-stranded form, it is also capable of forming duplexes in the same way as DNA; an example is an mRNA transcript pairing with an antisense version of the same transcript, which effectively silences the gene from which the mRNA was transcribed by preventing translation. As in dsDNA, the base pairing in dsRNA usually causes the twin strands to coil around each other in the shape of a double helix.
- downregulation
- Any process, natural or artificial, which decreases the level of gene expression of a certain gene. A gene which is observed to be expressed at relatively low levels (such as by detecting lower levels of its mRNA transcripts) in one sample compared to another sample is said to be downregulated. Contrast upregulation.
- downstream
- Towards or closer to the 3'-end of a chain of nucleotides, or to the C-terminus of a peptide chain. Contrast upstream.
- dsDNA
- See double-stranded DNA.
- dsRNA
- See double-stranded RNA.
- duplex
- See double-stranded.
- duplication
- The production of a second copy of part or all of a nucleotide sequence or amino acid sequence, either naturally or artificially, and the retention of both copies; especially when both the copy and the original sequence are retained in situ within the same molecule, often but not necessarily adjacent to each other. See also gene duplication, chromosomal duplication, and repeat.
- dyad
- See sister chromatids.
- dysplasia
- The abnormal growth or development of a tissue or organ; a change in the growth, behavior, or organization of cells within a tissue, or the presence of cells of an abnormal type, such that the tissue becomes disordered,[6] an event which often precedes the development of cancer.

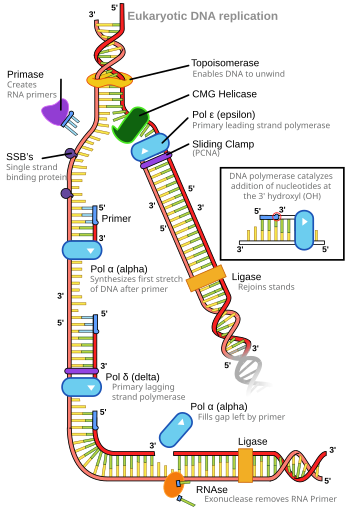

E
[edit]- eat-me signal
- A molecule exposed on the surface of a cell which effectively tags the cell for phagocytosis, inducing phagocytes to engulf or "eat" it. The presence of oxidized phospholipids or phosphatidylserine, or the absence of sialic acid from cell surface glycoproteins or glycolipids, are all commonly used as eat-me signals in certain cell types. See also find-me signal.
- electron transport chain
- electrophoresis
- The physical separation of molecules, e.g. nucleic acids or proteins, according to their movement through a fluid medium to which an electric field is applied. Because of their negatively charged phosphate backbones, nucleic acids are repelled by the negative electrode at one end of the medium and attracted to the positive electrode at the other end, which causes them to be pulled toward the latter over time; denatured proteins and even whole cells may migrate through the medium in a similar manner. The speed at which the molecules migrate depends on their net electric charge and is inversely proportional to their overall size (i.e. the number of atoms they contain), such that very small molecules tend to move faster through the medium than very large molecules. Thus electrophoretic techniques, particularly gel electrophoresis with agarose or polyacrylamide-based gels as the supporting medium, are widely used in molecular biology laboratories because they allow researchers to quickly and conveniently isolate molecules of interest from heterogeneous mixtures and/or identify them based on their expected molecular weight. Reference markers containing molecules of known molecular weight are commonly run alongside unknown samples to aid size-based identification. Electrophoresis is often combined with other techniques such as immunolabelling and radiolabelling.[8]
- electroporation
- A molecular biology technique in which a strong electric field is applied to living cells in order to temporarily increase the permeability of their cell membranes, allowing exogenous nucleic acids, proteins, or chemical compounds to easily pass through the membrane and thereby enter the cells. It is a common method of achieving transformation and transfection.
- elongation
- The linear growth of a nucleic acid polymer by the sequential addition of individual nucleotide monomers to a nascent strand, e.g. during transcription or replication, especially when it occurs by complementary pairing with a template strand. The term is often used to describe steps in certain laboratory techniques such as the polymerase chain reaction.
- elongation factor
- A protein which, by binding to a ribosome, promotes elongation of the polypeptide chain during translation.[8]
- embryo
- The developing organism that represents the earliest stages of development in all sexually reproducing multicellular organisms, traditionally encompassing the period after fertilization of an egg cell and formation of the zygote but prior to birth, hatching, or metamorphosis. During this period, known as embryonic development, the single-cell zygote is transformed by repeated cell divisions and rearrangements into a series of increasingly complex multicellular structures. For humans, the term "embryo" is only used until the ninth week after conception, after which time the embryo is known as a foetus; for most other organisms, including plants, "embryo" can be used more broadly to describe any early stage of the life cycle.
- emergenesis
- The quality of genetic traits that results from a specific configuration of interacting genes, rather than simply their combination.
- endocytosis
- Any process by which a substance is actively uptaken by or brought inside of a cell, crossing the plasma membrane from an extracellular space into an intracellular space, which includes the subclasses of pinocytosis, phagocytosis, and receptor-mediated processes. All of these involve surrounding an extracellular molecule, protein, or even another cell or organism with an extension or invagination of the cell membrane, which then "buds off" or separates from the rest of the membrane on the cytoplasmic side, forming a membrane-enclosed vesicle containing the ingested materials. By this mechanism the material can cross the lipid bilayer without being exposed to the hydrophobic space in between, instead remaining suspended in the fluid of the extracellular space. Many large, polar macromolecules which cannot simply diffuse across the membrane, such as metabolites and hormones, are transported into the cell by endocytosis. It is distinguished from alternative routes such as passing through protein channels or being chaperoned by transport proteins. The reverse process is called exocytosis.
- endomembrane
- Any membrane surrounding an intracellular organelle or vesicle, e.g. that of the endoplasmic reticulum, Golgi apparatus, lysosome, vacuole, nucleus (the nuclear envelope), etc.[8]
- endonuclease
- Any enzyme whose activity is to cleave phosphodiester bonds within a chain of nucleotides, including those that cleave relatively nonspecifically (without regard to sequence) and those that cleave only at very specific sequences (so-called restriction endonucleases). When recognition of a specific sequence is required, endonucleases make their cuts in the middle of the sequence. Contrast exonuclease.
- endoplasmic reticulum (ER)
- endosome
- enhancer
- A region of DNA near a gene that can be bound by an activator to increase gene expression or by a repressor to decrease expression.
- enhancer RNA (eRNA)
- A subclass of long non-coding RNAs transcribed from regions of DNA containing enhancer sequences. The expression of a given eRNA generally correlates with the activity of the corresponding enhancer in enhancing transcription of its target genes, suggesting that eRNAs play an active role in gene regulation in cis or in trans.
- enucleate
- To artificially remove the nucleus from a cell, e.g. by micromanipulation in the laboratory or by destroying it through irradiation with ultraviolet light, rendering the cell anucleate.[8]
- envagination
- enzyme
- A protein which acts as a catalyst for a biological process by accelerating a specific chemical reaction, typically by binding one or more substrate molecules and decreasing the activation energy necessary for the initiation of a particular reaction involving the substrate(s). Enzymatic catalysis often results in the chemical conversion of the substrate(s) into one or more products, which then inhibit or permit subsequent reactions. All metabolic pathways consist of a series of individual reactions which each depend upon one or more specific enzymes to drive them forward at rates fast enough to sustain life.
- epigenetics
- epigenome
- episome
- 1. Another name for a plasmid, especially one that is capable of integrating into a chromosome.
- 2. In eukaryotes, any non-integrated extrachromosomal circular DNA molecule that is stably maintained and replicated in the nucleus simultaneously with the rest of the host cell. Such molecules may include viral genomes, bacterial plasmids, and aberrant chromosomal fragments.
- epistasis
- The collective action of multiple genes interacting during gene expression. A form of gene action, epistasis can be either additive or multiplicative in its effects on specific phenotypic traits.
- epitope
- The specific site or region within an antigenic macromolecule such as a protein or carbohydrate which is recognized by B or T cells of the immune system, against which a specific antibody is produced, and with which the antibody's paratope specifically interacts or binds. In proteins, epitopes are typically motifs of 4–5 amino acid residues, sequential or discontiguous, which by virtue of the distinct spatial conformation they adopt upon protein folding are able to uniquely interact with a particular paratope. In this sense they may be considered binding sites, though they do not necessarily overlap with ligand binding sites and need not be in any way relevant to the protein's normal function. Very large molecules may have multiple epitopes, each of which is recognized by a different antibody.
- ergosome
- See polysome.
- euchromatin
- A relatively open, lightly compacted form of chromatin in which DNA is only sporadically bound in nucleosomes and thus broadly accessible to binding and manipulation by proteins and other molecules. Euchromatic regions of a genome are often enriched in genes and actively undergoing transcription, in contrast to heterochromatin, which is relatively gene-poor, nucleosome-rich, and less accessible to transcription machinery.
- euploidy
- The condition of a cell or organism having an abnormal number of complete sets of chromosomes, possibly excluding the sex chromosomes. Euploidy differs from aneuploidy, in which a cell or organism has an abnormal number of one or more specific individual chromosomes.
- evolution
- The change in the heritable characteristics of biological populations over successive generations. In the most traditional sense, it occurs by changes in the frequencies of alleles in a population's gene pool.
- ex vivo
- Occurring outside of a cell or organism, as with observations made or experiments performed in or on cells or tissues which have been isolated or removed from their natural context to an external environment (usually a carefully controlled environment with minimal alteration of natural conditions, such as a cell culture being grown in a laboratory). This is in contrast to in vivo observations, which are made in an entirely natural context.
- excision
- The enzymatic removal of a polynucleotide sequence from one or more strands of a nucleic acid, or of a polypeptide sequence from a protein, typically implying both the breaking of the polymeric molecule in two locations and the subsequent rejoining of the two breakpoints after the sequence between them has been removed. The term may be used to describe a wide variety of processes performed by distinct enzymes, including most splicing and DNA repair pathways.[4]
- exocytosis
- Any active transport process by which a substance is secreted from or transported out of a cell, crossing the plasma membrane from the interior of the cell into the extracellular space, especially that which occurs by the fusion of the membrane surrounding a secretory vesicle with the larger cell membrane. This fusion causes the intra-vesicular space to merge with the extracellular fluid, releasing the vesicle's contents on the exterior side of the cell without exposing them to the hydrophobic space between the lipid bilayer. More narrowly the term may refer in particular to the bulk transport of a large amount of molecules out of the cell all at once, often metabolites or hormones which are too large and polar to passively diffuse across the membrane themselves. The reverse process, whereby materials are invaginated into the cell, is known as endocytosis.
- exome
- The entire set of exons within a particular genome, including untranslated regions of mature mRNAs as well as coding regions.
- exon
- Any part of a gene that encodes a part of the final mature messenger RNA produced by that gene after introns have been removed by alternative splicing. The term refers to both the sequence as it exists within a DNA molecule and to the corresponding sequence in RNA transcripts.
- exon skipping
- exonuclease
- Any enzyme whose activity is to cleave phosphodiester bonds within a chain of nucleotides, including those that cleave only upon recognition of a specific sequence (so-called restriction exonucleases). Exonucleases make their cuts at either the 3' or 5'-end of the sequence (rather than in the middle, as with endonucleases).
- exosome
- 1. (protein complex) An intracellular multi-protein complex which serves the function of degrading various types of RNA molecules.
- 2. (vesicle) A type of membrane-bound extracellular vesicle produced in many eukaryotic cells by the inward budding of an endosome and the subsequent fusion of the endosome with the plasma membrane, causing the release of the vesicle into various extracellular spaces, including biological fluids such as blood and saliva, where they may serve any of a wide variety of physiological functions, from waste management to intercellular signaling.
- exosome complex
- An intracellular multi-protein complex which serves the function of degrading various types of RNA molecules.
- expression vector
- A type of vector, usually a plasmid or viral vector, designed specifically for the expression of a transgene insert in a target cell, rather than for some other purpose such as cloning.
- extein
- Any part of an amino acid sequence which is retained within a precursor polypeptide, i.e. not excised by post-translational protein splicing, and is therefore present in the mature protein, analogous to the exons of RNA transcripts. Contrast intein.
- extension
- See elongation.
- extracellular
- Outside the plasma membrane of a cell or cells; i.e. located or occurring externally to a cell. Contrast intracellular; see also intercellular.
- extracellular matrix (ECM)
- The network of interacting macromolecules and minerals secreted by and existing outside of and between cells in multicellular structures such as tissues and biofilms, forming a hydrated, mesh-like, semi-solid suspension which not only holds the cells together in an organized fashion but also provides structural and biochemical support, acting as an elastic, compressible buffer against external stresses as well as both regulating and influencing numerous aspects of cell behavior, among them cell adhesion, motility, metabolism, division, and cell-to-cell communication. The composition and properties of the ECM vary enormously between organisms and tissue types, but generally it takes the form of a polysaccharide gel in which various fibrous proteins (especially collagen and elastin), enzymes, and glycoproteins are embedded. Cells themselves both produce the matrix components and respond constantly to local matrix composition, a source of environmental feedback which is critical for differentiation, tissue organization, and development.[8][21]
- extrachromosomal DNA
- Any DNA that is not found in chromosomes or in the nucleus of a cell and hence is not genomic DNA. This may include the DNA contained in plasmids or organelles such as mitochondria or chloroplasts, or, in the broadest sense, DNA introduced by viral infection. Extrachromosomal DNA usually shows significant structural differences from nuclear DNA in the same organism.
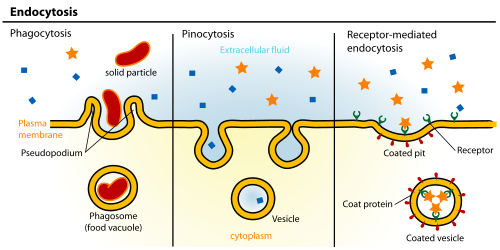
F
[edit]- facilitated diffusion
- A type of passive transport by which substances are conveyed across membranes more quickly than would be possible by ordinary passive diffusion alone, generally because proteins embedded within the membrane act as shuttles or pores, being arranged in such a way as to provide a hydrophilic environment that is favorable for the movement of small polar molecules, which would otherwise be repulsed by the hydrophobic interior of the lipid bilayer.[12]
- facultative expression
- The transcription of a gene only as needed, as opposed to constitutive expression, in which a gene is transcribed continuously. A gene that is transcribed as needed is called a facultative gene.
- fatty acid
- fermentation
- filopodium
- find-me signal
- A molecule exposed on the surface of a cell destined for apoptosis which is used to attract phagocytes to engulf and eliminate the cell by phagocytosis. See also eat-me signal.
- five-prime cap
- See 5' cap.
- five-prime end
- See 5'-end.
- five-prime untranslated region
- See 5' untranslated region.
- fixation
- 1. (histology) The preservation of biological material by treating it with a chemical fixative that prevents or delays the natural postmortem processes of decay (e.g. autolysis and putrefaction) which would otherwise eventually cause cells, tissues, and biomolecules to lose their characteristic structures and properties. Biological specimens are usually fixed with the broad objective of arresting or slowing biochemical reactions for long enough to study them in detail, essentially 'freezing' cellular processes in their natural state at a specific point in time, while minimizing disruption to existing structures and arrangements, all of which can improve subsequent staining and microscopy of the fixed samples. Though fixation tends to irreversibly terminate any ongoing reactions, thus killing the fixed cells, it makes it possible to study molecular details that occur too rapidly or transiently to observe in living samples. Common fixatives such as formaldehyde work by disabling proteolytic enzymes, coagulating, insolubilizing, and/or denaturing macromolecules, creating crosslinks between them, and protecting specimens from decomposition by bacteria and fungi.
- 2. (population genetics) The process by which a single allele for a particular gene with multiple different alleles increases in frequency in a given population such that it becomes permanently established as the only allele at that locus within the population's gene pool.
- fixative
- Any chemical compound or solution that causes the fixation of cells, tissues, or other microscopic structures by any mechanism, thus preserving them for long-term, detailed study by methods such as embedding, staining, and microscopy. Common fixatives include dilute solutions of ethanol, acetic acid, formaldehyde, and osmium tetroxide, among others.[8]
- flagellate
- (of a cell) Having one or more flagella.
- flagellum
- A long, thin, hair-like appendage protruding from the surface of some cells, which serves locomotory functions by undulating in a way that propels the cell through its environment or by effecting the movement of extracellular fluids and solutes past the cell surface. Many unicellular organisms, including some bacteria, protozoa, and algae, bear one or more flagella, and certain cell types in multicellular organisms, namely sperm cells, also have flagella. Eukaryotic flagella are essentially just longer versions of cilia, often up to 150 micrometres (μm) in length, while bacterial flagella are typically smaller and completely different in structure and mechanism of action.[6][8]
- fluorescence in situ hybridization (FISH)
- forward genetics
- An experimental approach in molecular genetics in which a researcher starts with a specific known phenotype and attempts to determine the genetic basis of that phenotype by any of a variety of laboratory techniques, commonly by inducing random mutations in the organism's genome and then screening for changes in the phenotype of interest. Observed phenotypic changes are assumed to have resulted from the mutation(s) present in the screened sample, which can then be mapped to specific genomic loci and ultimately to one or more specific candidate genes. This methodology contrasts with reverse genetics, in which a specific gene or its gene product is individually manipulated in order to identify the gene's function.
- forward mutation
- frameshift mutation
- A type of mutation in a nucleic acid sequence caused by the insertion or deletion of a number of nucleotides that is not divisible by three. Because of the triplet nature by which nucleotides code for amino acids, a mutation of this sort causes a shift in the reading frame of the nucleotide sequence, resulting in the sequence of codons downstream of the mutation site being completely different from the original.
- freeze-drying
- See lyophilization.
- Functional Genomics Data (FGED) Society
- An organization that works with others "to develop standards for biological research data quality, annotation and exchange" as well as software tools that facilitate their use.[22]
G
[edit]- G banding
- A technique used in cytogenetics to produce a visible karyotype by staining the condensed chromosomes with Giemsa stain. The staining produces consistent and identifiable patterns of dark and light "bands" in regions of chromatin, which allows specific chromosomes to be easily distinguished.
- G1
- G2
- gamete
- A haploid cell that is the meiotic product of a progenitor germ cell and the final product of the germ line in sexually reproducing multicellular organisms. Gametes are the means by which an organism passes its genetic information to its offspring; during fertilization, two gametes (one from each parent) are fused into a single diploid zygote.
- gametogenesis
- The process by which eukaryotic precursor germ cells divide and differentiate into haploid gametes. Depending on the organism, gametes may be generated from haploid germ cells by mitosis or diploid germ cells by meiosis.
- GC content
- See guanine-cytosine content.
- gDNA
- See genomic DNA.
- gene
- Any segment or set of segments of a nucleic acid molecule that contains the information necessary to produce a functional RNA transcript in a controlled manner. In living organisms, genes are often considered the fundamental units of heredity and are typically encoded in DNA. A particular gene can have multiple different versions, or alleles, and a single gene can result in a gene product that influences many different phenotypes.
- gene cassette
- See cassette.
- gene dosage
- The number of copies of a particular gene present in a genome. Gene dosage directly influences the amount of gene product a cell is able to express, though a variety of controls have evolved which tightly regulate gene expression. Changes in gene dosage caused by mutations include copy-number variations.
- gene duplication
- A type of mutation defined as any duplication of a region of DNA that contains a gene. Compare chromosomal duplication.
- gene expression
- The set of processes by which the information encoded in a gene is used in the synthesis of a gene product, such as a protein or non-coding RNA, or otherwise made available to influence one or more phenotypes. Canonically, the first step is transcription, which produces a messenger RNA molecule complementary to the DNA molecule in which the gene is encoded; for protein-coding genes, the second step is translation, in which the messenger RNA is read by the ribosome to produce a protein. The information contained within a DNA sequence need not necessarily be transcribed and translated to exert an influence on molecular events, however; broader definitions encompass a huge variety of other ways in which genetic information can be expressed.
- Gene Expression Omnibus (GEO)
- A database of high-throughput functional genomics and gene expression data derived from experimental chips and next-generation sequencing and managed by the National Center for Biotechnology Information.[23][24]
- gene fusion
- The union, either by natural mutation or by recombinant laboratory techniques, of two or more previously independent genes that code for different gene products such that they become subject to control by the same regulatory systems. The resulting hybrid sequence is known as a fusion gene.[4]
- gene mapping
- Any of a variety of methods used to precisely identify the location of a particular gene within a DNA molecule (such as a chromosome) and/or the physical or linkage distances between it and other genes.
- gene of interest (GOI)
- A gene being studied in a scientific experiment, especially one that is the focus of a genetic engineering technique such as cloning.
- gene product
- Any of the biochemical material resulting from the expression of a gene, most commonly interpreted as the functional mRNA transcript produced by transcription of the gene or the fully constructed protein produced by translation of the transcript, though non-coding RNA molecules such as transfer RNAs may also be considered gene products. A measurement of the quantity of a given gene product that is detectable in a cell or tissue is sometimes used to infer how active the corresponding gene is.
- gene regulation
- The broad range of mechanisms used by cells to control the activity of their genes, especially to allow, prohibit, increase, or decrease the production or expression of specific gene products, such as RNA or proteins. Gene regulation increases an organism's versatility and adaptability by allowing its cells to express different gene products when required by changes in its environment. In multicellular organisms, the regulation of gene expression also drives cellular differentiation and morphogenesis in the embryo, enabling the creation of a diverse array of cell types from the same genome.
- gene silencing
- Any mechanism of gene regulation which drastically reduces or completely prevents the expression of a particular gene. Gene silencing may occur naturally during either transcription or translation. Laboratory techniques often exploit natural silencing mechanisms to achieve gene knockdown.
- gene therapy
- The insertion of a functional or wild-type gene or part of a gene into an organism (especially a patient) with the intention of correcting a genetic defect, either by direct substitution of the defective gene or by supplementation with a second, functional version.[14]
- gene trapping
- A high-throughput technology used to simultaneously inactivate, identify, and report the expression of a target gene in a mammalian genome by introducing an insertional mutation consisting of a promoterless reporter gene and/or a selectable marker flanked by an upstream splice site and a downstream polyadenylated termination sequence.
- generation
- 1. In any given organism, a single reproductive cycle, or the phase between two consecutive reproductive events, i.e. between an individual organism's reproduction and that of the progeny of that reproduction; or the actual or average length of time required to complete a single reproductive cycle, either for a particular lineage or for a population or species as a whole.
- 2. In a given population, those individuals (often but not necessarily living contemporaneously) who are equally removed from a given common ancestor by virtue of the same number of reproductive events having occurred between them and the ancestor.[14]
- genetic background
- genetic code
- A set of rules by which information encoded within nucleic acids is translated into proteins by living cells. These rules define how sequences of nucleotide triplets called codons specify which amino acid will be added next during protein synthesis. The vast majority of living organisms use the same genetic code (sometimes referred to as the "standard" genetic code) but variant codes do exist.
- genetic disorder
- Any illness, disease, or other health problem directly caused by one or more abnormalities in an organism's genome which are congenital (present at birth) and not acquired later in life. Causes may include a mutation to one or more genes, or a chromosomal abnormality such as an aneuploidy of a particular chromosome. The mutation responsible may occur spontaneously during embryonic development or may be inherited from one or both parents, in which case the genetic disorder is also classified as a hereditary disorder. Though the abnormality itself is present before birth, the actual disease it causes may not develop until much later in life; some genetic disorders do not necessarily guarantee eventual disease but simply increase the risk of developing it.
- genetic distance
- A measure of the genetic divergence between species, populations within a species, or individuals, used especially in phylogenetics to express either the time elapsed since the existence of a common ancestor or the degree of differentiation in the DNA sequences comprising the genomes of each population or individual.
- genetic engineering
- The direct, deliberate manipulation of an organism's genetic material using any of a variety of biotechnology methods, including the insertion or removal of genes, the transfer of genes within and between species, the mutation of existing sequences, and the construction of novel sequences using artificial gene synthesis. Genetic engineering encompasses a broad set of technologies by which the genetic composition of individual cells, tissues, or entire organisms may be altered for various purposes, commonly in order to study the functions and expression of individual genes, to produce hormones, vaccines, and other drugs, and to create genetically modified organisms for use in research and agriculture.
- genetic marker
- A specific, easily identifiable, and usually highly polymorphic gene or other DNA sequence with a known location on a chromosome that can be used to identify the individual or species possessing it.
- genetic recombination
- Any reassortment or exchange of genetic material within an individual organism or between individuals of the same or different species, especially that which creates genetic variation. In the broadest sense, the term encompasses a diverse class of naturally occurring mechanisms by which nucleic acid sequences are copied or physically transferred into different genetic environments, including homologous recombination during meiosis or mitosis or as a normal part of DNA repair; horizontal gene transfer events such as bacterial conjugation, viral transduction, or transformation; or errors in DNA replication or cell division. Artificial recombination is central to many genetic engineering techniques which produce recombinant DNA.
- genetic redundancy
- The redundant encoding of two or more distinct gene products that ultimately perform the same biochemical function. Mutations in one of these genes may have a smaller effect on fitness than might be expected, since the redundant genes often compensate for any loss of function and obviate any gain of function.
- genetic regulatory network (GRN)
- A graph that represents the regulatory complexity of gene expression. The vertices (nodes) are represented by various regulatory elements and gene products while the edges (links) are represented by their interactions. These network structures also represent functional relationships by approximating the rate at which genes are transcribed.
- genetic testing
- A broad class of various procedures used to identify features of an individual's particular chromosomes, genes, or proteins in order to determine parentage or ancestry, diagnose vulnerabilities to heritable diseases, or detect mutant alleles associated with increased risks of developing genetic disorders. Genetic testing is widely used in human medicine, agriculture, and biological research.
- genetically modified organism (GMO)
- Any organism whose genetic material has been altered using genetic engineering techniques, particularly in a way that does not occur naturally by mating or by natural genetic recombination.
- genetics
- The field of biology that studies genes, genetic variation, and heredity in living organisms.
- genome
- 1. The entire complement of genetic material contained within the chromosomes of an organism, organelle, or virus.
- 2. The collective set of genes or genetic loci shared by every member of a population or species, regardless of the different alleles that may be present at these loci in different individuals.
- genome instability
- genome size
- The total amount of DNA contained within one copy of a genome, typically measured by mass (in picograms or daltons) or by the total number of base pairs (in kilobases or megabases). For diploid organisms, genome size is often used interchangeably with C-value.
- genome-wide association study (GWAS)
- genomic DNA (gDNA)
- The DNA contained in chromosomes, as opposed to the extrachromosomal DNA contained in separate structures such as plasmids or organelles such as mitochondria or chloroplasts.
- genomic imprinting
- An epigenetic phenomenon that causes genes to be expressed in a manner dependent upon the particular parent from which the gene was inherited. It occurs when epigenetic marks such as DNA or histone methylation are established or "imprinted" in the germ cells of a parent organism and subsequently maintained through cell divisions in the somatic cells of the organism's progeny; as a result, a gene in the progeny that was inherited from the father may be expressed differently than another copy of the same gene that was inherited from the mother.
- genomic island (GI)
- A region of a genome that shows evidence of horizontal transfer from another organism. The term is used especially in describing microbial genomes such as those of bacteria, where genomic islands having the same or similar sequences commonly occur in species or strains that are otherwise only distantly related, implying that they were not passed on through vertical descent from a common ancestor but through some form of lateral transfer such as conjugation. These islands often contain functional genes which confer adaptive traits such as antibiotic resistance.
- genomics
- An interdisciplinary field that studies the structure, function, evolution, mapping, and editing of entire genomes, as opposed to individual genes.
- genotoxicity
- The ability of certain chemical agents to cause damage to genetic material within a living cell (e.g. through single- or double-stranded breaks, crosslinking, or point mutations), which may or may not result in a permanent mutation. Though all mutagens are genotoxic, not all genotoxic compounds are mutagenic.
- genotype
- The entire complement of alleles present in a particular individual's genome, which gives rise to the individual's phenotype.
- genotyping
- The process of determining differences in the genotype of an individual by examining the DNA sequences in the individual's genome using bioassays and comparing them to another individual's sequences or a reference sequence.
- germ cell
- Any cell that gives rise to the gametes of a sexually reproducing organism. Germ cells are the vessels for the genetic material which will ultimately be passed on to the organism's descendants and are usually distinguished from somatic cells, which are entirely separate from the germ line.
- germ line
- 1. In multicellular organisms, the subpopulation of cells which are capable of passing on their genetic material to the organism's progeny and are therefore (at least theoretically) distinct from somatic cells, which cannot pass on their genetic material except to their own immediate mitotic daughter cells. Cells of the germ line are called germ cells.
- 2. The lineage of germ cells, spanning many generations, that contains the genetic material which has been passed on to an individual from its ancestors.
- gigabase (Gb)
- A unit of nucleic acid length equal to one billion (1×109) bases in single-stranded molecules or one billion base pairs in duplex molecules such as double-stranded DNA.
- glucogenic amino acid
- Any amino acid that can be converted into glucose via gluconeogenesis, as opposed to the ketogenic amino acids, which can be converted into ketone bodies. In humans, 18 of the 20 amino acids are glucogenic; only leucine and lysine are not. Five amino acids (phenylalanine, isoleucine, threonine, tryptophan, and tyrosine) are both glucogenic and ketogenic.
- gluconeogenesis (GNG)
- The chain of metabolic reactions that results in the generation of glucose from some non-carbohydrate carbon substrates, including the glucogenic amino acids. It is one of two primary pathways used by most animals to maintain blood sugar levels (the other being glycogenolysis), especially during periods of fasting, starvation, and intense exercise.
- glucose
- A simple sugar with the molecular formula C
6H
12O
6 and the most abundant monosaccharide in nature, being the primary product of photosynthesis, where it is made in a sunlight-powered reaction of water with carbon dioxide. All living organisms are capable of metabolizing glucose via glycolysis, an exergonic pathway which for most organisms is the primary means of obtaining chemical energy to power cellular activities.[8] Metabolic glucose is usually stored in the form of large polymeric aggregates such as amylose in plants and glycogen in animals, and is released by the breakdown of these polymers via glycogenolysis. - glycogen
- A branched polysaccharide composed of as many as 30,000 covalently bonded units of the monosaccharide glucose which functions as the primary form of short-term energy storage in most animal cells.[8][12] Glycogen reserves are especially abundant in muscle and liver cells,[6] where they can be metabolized at-need into their component glucoses as a means of buffering blood sugar levels, a process known as glycogenolysis.
- glycogenolysis
- A metabolic pathway in which polymeric glycogen molecules are broken down into individual glucose monomers by the sequential removal of glucose units via phosphorolysis, a reaction catalyzed by the enzyme glycogen phosphorylase. Glycogenolysis is one of two primary pathways used in animal tissues to generate free glucose for the maintenance of blood sugar levels, the other being gluconeogenesis.
- glycolipid
- Any of a subclass of lipids consisting of a central polar molecule (most commonly glycerol or sphingosine) which is covalently attached to one or more monosaccharides or oligosaccharides via glycosidic bonds, as well as to one or more long, non-polar fatty acid chains.[6] Glycolipids are one of three major types of membrane lipid comprising all biological membranes, along with phospholipids and cholesterol.
- glycolysis
- The metabolic pathway in which carbohydrate sugars such as glucose are broken down into simpler molecules, releasing chemical energy which can then be used for various cellular functions. In a series of ten enzyme-catalyzed reactions, each molecule of glucose is converted into two molecules of pyruvate, with the free energy liberated in this process simultaneously being used to form high-energy bonds in two molecules of reduced nicotinamide adenine dinucleotide (NADH) and two molecules of adenosine triphosphate (ATP). In aerobic conditions pyruvate and NADH are further oxidized in the mitochondria; in anaerobic conditions NADH itself subsequently reduces pyruvate to lactate.
- glycoprotein
- A protein with one or more carbohydrate molecules, typically short oligosaccharide chains, covalently attached to one or more of its amino acid side chains.[6] Proteins which are exposed on the outer surface of the plasma membrane or are secreted into the extracellular space are commonly glycosylated in this way.
- glycosidase
- Any of a class of enzymes capable of breaking one or more glycosidic bonds in carbohydrate molecules, commonly found in lysosomes.[8]
- glycoside
- Any chemical compound in which a carbohydrate molecule is covalently bonded to another molecule containing a hydroxyl group (including other carbohydrates) via one or more C–O glycosidic bonds. When both molecules are carbohydrates, the glycoside is a disaccharide or polysaccharide.[8]
- glycosidic bond
- A covalent ether bond that connects a carbon atom within a carbohydrate molecule (e.g. a monosaccharide) or a carbohydrate derivative to another substituent or functional group, which may or may not be another carbohydrate; such bonds form as the result of a dehydration reaction between hydroxyl groups on each molecule, liberating a water molecule in the process. A substance containing a glycosidic bond is known as a glycoside.
- glycosylation
- The attachment of an oligosaccharide (e.g. glucose) to an asparagine residue within a peptide or protein by covalent bonding, a process which takes place in or near the rough endoplasmic reticulum.[8]
- Goldberg-Hogness box
- See TATA box.
- Golgi apparatus
- gRNA
- See guide RNA.
- guanine (G)
- A purine nucleobase used as one of the four standard nucleobases in both DNA and RNA molecules. Guanine forms a base pair with cytosine.
- guanine-cytosine content
- The proportion of nitrogenous bases in a nucleic acid that are either guanine (G) or cytosine (C), typically expressed as a percentage. DNA and RNA molecules with higher GC-content are generally more thermostable than those with lower GC-content due to molecular interactions that occur during base stacking.[25]
- guanosine (G, Guo)
- One of the four standard nucleosides used in RNA molecules, consisting of a guanine base with its N9 nitrogen bonded to the C1 carbon of a ribose sugar. Guanine bonded to deoxyribose is known as deoxyguanosine, which is the version used in DNA.
- guide RNA (gRNA)
- A short single-stranded RNA oligonucleotide which complexes with Cas endonucleases and, by annealing to a specific complementary sequence in a DNA molecule, serves to "guide" these proteins to viral DNA introduced by foreign pathogens, which can then be digested and degraded as part of an adaptive immune defense employed by bacteria and archaea. Custom-made guide RNAs are designed by scientists to target specific genomic loci in CRISPR-Cas gene editing.
H
[edit]- hairpin
- See stem-loop.
- haploid
- (of a cell or organism) Having one copy of each chromosome, with each copy not being part of a pair. Contrast diploid and polyploid.
- Hayflick limit
- helicase
- Any of a class of ATP-dependent motor proteins that move directionally along the DNA backbone and catalyze the separation of the two complementary strands of double-stranded molecules, permitting a wide variety of vital processes to take place, e.g. transcription, replication, and repair.[14]
- hemizygous
- In a diploid organism, having just one allele at a given genetic locus (where there would ordinarily be two). Hemizygosity may be observed when only one copy of a chromosome is present in a normally diploid cell or organism, or when a segment of a chromosome containing one copy of an allele is deleted, or when a gene is located on a sex chromosome in the heterogametic sex (in which the sex chromosomes do not exist in matching pairs); for example, in human males with normal chromosomes, almost all X-linked genes are said to be hemizygous because there is only one X chromosome and few of the same genes exist on the Y chromosome.
- heredity
- The storage, transfer, and expression of molecular information in biological organisms,[14] as manifested by the passing on of phenotypic traits from parents to their offspring, either through sexual or asexual reproduction. Offspring cells or organisms are said to inherit the genetic information of their parents.
- heterochromatin
- heterochromosome
- See allosome.
- heterodimer
- A protein or protein domain composed of two different polypeptides which are paired with each other in the quaternary structure of a multimeric complex.[4] Contrast homodimer.
- heterogeneous nuclear RNA (hnRNA)
- heterokaryon
- A multinucleate cell containing nuclei with different genotypes, resulting from the fusion of two or more genetically distinct cells, either naturally (e.g. in certain types of sexual reproduction) or artificially (e.g. in genetic engineering).[8]
- heterologous expression
- The expression of a foreign gene or any other foreign DNA sequence within a host organism which does not naturally contain the same gene. Insertion of foreign transgenes into heterologous hosts using recombinant vectors is a common biotechnology method for studying gene structure and function.
- heterozygous
- In a diploid organism, having two different alleles at a given genetic locus. In genetics shorthand, heterozygous genotypes are represented by a pair of non-matching letters or symbols, often an uppercase letter (indicating a dominant allele) and a lowercase letter (indicating a recessive allele), such as "Aa" or "Bb". Contrast homozygous.
- high-throughput
- histology
- The study or analysis of the microscopic anatomy of biological tissues or of cells within tissues, particularly by making use of specialized techniques to distinguish structures and functions based on visual morphology and differential staining. In practice the term is sometimes used more broadly to include cytology.
- histone
- Any of a class of highly alkaline proteins responsible for packaging nuclear DNA into structural units called nucleosomes in eukaryotic cells. Histones are the chief protein components of chromatin, where they associate into eight-membered complexes which act as "spools" around which the linear DNA molecule winds. They play a major role in gene regulation and expression.
- histone core
- The complex of eight histone proteins around which double-stranded DNA wraps within a nucleosome. The canonical histone octamer consists of two each of histones H2A, H2B, H3, and H4, which pair with each other symmetrically to form a ball-shaped cluster around which DNA winds through interactions with the histones' surface domains, though variant histones may replace their analogues in certain contexts.
- histone modification
- histone variant
- hnRNA
- See heterogeneous nuclear RNA.
- holocentric
- (of a linear chromosome or chromosome fragment) Having no single centromere but rather multiple kinetochore assembly sites dispersed along the entire length of the chromosome. During cell division, the chromatids of holocentric chromosomes move apart in parallel and do not form the classical V-shaped structures typical of monocentric chromosomes.
- homeobox
- Any of a class of DNA sequences approximately 180 base pairs in length occurring near the 3'‐end of certain eukaryotic genes and encoding a 60-amino acid domain, known as a homeodomain, which is capable of binding to DNA or RNA via a characteristic helix-turn-helix motif. Homeobox-containing genes are translated into homeodomain-containing proteins, which commonly regulate transcription or translation by binding to other genes or messenger RNAs containing homeobox responsive elements. The products of many homeotic genes, exemplified by the Hox genes, are of critical importance in developmental pathways.[4]
- homeobox responsive element (HRE)
- Any DNA or RNA sequence that is specifically recognized and bound by the homeodomain of a homeodomain-containing protein.
- homeodomain
- A nucleic acid-binding domain, typically 60 amino acids in length, found near the C-terminus of certain eukaryotic proteins, characterized by a highly conserved helix-turn-helix motif that binds with strong affinity to the backbone of specific recognition sequences in DNA or RNA molecules. A protein may have one or more homeodomains, each of which is specific to a different recognition sequence. Many homeodomain-containing proteins function as transcription factors by binding to sequences within promoters and blocking or recruiting other proteins, such as RNA polymerase or cofactors of the transcription initiation complex. Homeodomains are the translated versions of homeoboxes, though the terms are often used interchangeably.
- homodimer
- A protein or protein domain composed of two identical polypeptides which are paired with each other in the quaternary structure of a multimeric complex.[4] Contrast heterodimer.
- homologous chromosomes
- A set of two matching chromosomes, one maternal and one paternal, which pair up with each other inside the nucleus during meiosis. They have the same genes at the same loci, but may have different alleles.
- homologous recombination
- A type of genetic recombination in which nucleotide sequences are exchanged between two similar or identical ("homologous") molecules of DNA, especially that which occurs between homologous chromosomes. The term may refer to the recombination that occurs as a part of any of a number of distinct cellular processes, most commonly DNA repair or chromosomal crossover during meiosis in eukaryotes and horizontal gene transfer in prokaryotes. Contrast nonhomologous recombination.
- homozygous
- In a diploid organism, having two identical alleles at a given genetic locus. In genetics shorthand, homozygous genotypes are represented by a pair of matching letters or symbols, such as "AA" or "aa". Contrast heterozygous.
- horizontal gene transfer (HGT)
- Any process by which genetic material is transferred between unicellular and/or multicellular organisms other than by vertical transmission from parent to offspring, e.g. bacterial conjugation.
- housekeeping gene
- Any constitutive gene that is transcribed at a relatively constant level across many or all known conditions and cell types. The products of housekeeping genes typically play critical roles in the maintenance of cellular integrity and basic metabolic function. It is generally assumed that their expression is unaffected by experimental or pathological conditions.
- Hox genes
- A subset of highly conserved homeobox-containing genes whose protein products function as transcription factors essential for the proper organization of the body plan in developing animal embryos, ensuring that the correct structures are formed in the correct places. Hox genes are usually arranged on a chromosome in tandem arrays and are expressed sequentially during development, with the sequence of gene activation corresponding to their physical arrangement within the genome and/or the physical layout of the tissues in which they are expressed along the organism's anterior–posterior axis.[4]
- Human Genome Project (HGP)
- A collaborative international scientific research project with the goal of sequencing all of the chromosomal DNA and identifying and mapping all of the genes within human cells, and ultimately of assembling a complete reference genome for the human species. The project was launched in 1990 by a consortium of federal agencies, universities, and research institutions and was declared complete in 2003. Because each individual human being has a unique genome, the finished reference genome is a mosaic of sequences obtained by sampling DNA from thousands of individuals across the world and does not represent any one individual.
- hyaloplasm
- See cytosol.
- hybrid
- The offspring that results from combining the qualities of two organisms of different genera, species, breeds, or varieties through sexual reproduction. Hybrids may occur naturally or artificially, as during selective breeding of domesticated animals and plants. Reproductive barriers typically prevent hybridization between distantly related organisms, or at least ensure that hybrid offspring are sterile, but fertile hybrids may result in speciation.
- hybridization
- 1. The process by which a hybrid organism is produced from two organisms of different genera, species, breeds, or varieties.
- 2. The process by which two or more single-stranded nucleic acid molecules with complementary nucleotide sequences pair with each other in solution, creating double-stranded or triple-stranded molecules via the formation of hydrogen bonds between the complementary nucleobases of each strand. In certain laboratory contexts, especially ones in which long strands hybridize with short oligonucleotide primers, hybridization is often referred to as annealing.
- 3. A step in some experimental assays in which a single-stranded DNA or RNA preparation is added to an array surface and anneals to a complementary hybridization probe.
- hybridization probe
- hydrophilic
- Soluble in or having an affinity for water or other polar compounds; describing a polar molecule, or a moiety or functional group within a molecule, which participates in intermolecular interactions such as hydrogen bonding with other polar molecules and therefore readily dissolves in polar solvents such as water or aqueous solutions.[6] Unlike hydrophobic compounds, hydrophilic compounds can form energetically favorable contacts with the aqueous phase of biological fluids and so can often be suspended directly in the cytosol or exposed to extracellular spaces.[12] Together, the contrasting properties of hydrophilicity and hydrophobicity play major roles in determining the structural conformations and functions of most biomolecules.
- hydrophobic
- Having a low solubility in or affinity for water or other polar solvents; describing a non-polar molecule, or a moiety or functional group within a molecule, which cannot form energetically favorable interactions with polar compounds and which therefore tends to "avoid" or be repulsed by such compounds, instead clustering together with other hydrophobic molecules or arranging itself in a way that minimizes its exposure to its polar surroundings. This phenomenon is not so much due to the affinity of the hydrophobic molecules for each other as it is a consequence of the strong intermolecular forces that allow polar compounds such as water molecules to bond with each other; hydrophobic species are unable to form alternative bonds of equivalent strength with the polar compounds, hence they tend to be excluded from aqueous solutions by the tendency of the polar solvent to maximize interactions with itself. Hydrophobicity is a major determinant of countless chemical interactions in biological systems, including the spatial conformations assumed by macromolecules such as proteins and lipids, the binding of ligands and substrates to proteins, and the structure and properties of lipid membranes.[8][6] Contrast hydrophilic.
- hypertonic
- Describing a solution containing a high concentration of dissolved solutes relative to another solution, i.e. having positive osmotic pressure, such that solvent will tend to move by osmosis across a semipermeable membrane from the solution of lower solute concentration to the solution of higher concentration until both solutions have equal concentrations. In a cell where the intracellular cytosol is hypertonic relative to the surrounding extracellular fluid (which by definition is hypotonic relative to the cytosol), the solvent (water) will flow across the plasma membrane into the cytosol, filling the cell with extra water and diluting its contents until both sides of the membrane are isotonic. Cells placed in severely hypotonic environments may be at risk of bursting due to the sudden inflow.
- hypomorph
- A mutant allele that permits a subnormal expression of the gene's normal phenotype, e.g. by encoding an unstable enzyme which degrades too quickly to fully serve its function but which nevertheless is functional in some limited capacity, being generated in quantities sufficient for its reaction to proceed slowly or at low levels.[4]
- hypotonic
- Describing a solution containing a low concentration of dissolved solutes relative to another solution, i.e. having negative osmotic pressure, such that solvent will tend to move by osmosis across a semipermeable membrane from the solution of lower solute concentration to the solution of higher concentration until both solutions have equal concentrations. In a cell where the intracellular cytosol is hypotonic relative to the surrounding extracellular fluid (which by definition is hypertonic relative to the cytosol), the solvent (water) will flow across the plasma membrane out of the cytosol, causing the cell to lose water until both sides of the membrane are isotonic. Cells placed in severely hypertonic environments may be at risk of shriveling and desiccating due to the sudden outflow.
- hypoxanthine (I)
- A naturally occurring non-canonical purine nucleobase that is used in some RNA molecules and pairs with standard nucleobases in a phenomenon known as wobble base pairing. Its nucleoside form is known as inosine, which is the reason it is commonly abbreviated with the letter I in sequence reads.
I
[edit]- idiochromosome
- See allosome.
- idiogram
- A diagrammatic or schematic karyotype of the entire set of chromosomes within a cell or genome, in which annotated illustrations depict each chromosome in its most idealized form (e.g. with straight lines and obvious centromeres) so as to facilitate the easy identification of sequences, structural features, and physical distances, which may be less apparent in photomicrographs of the actual chromosomes.
- idiomere
- See chromomere.
- immortalization
- immunogenic
- Capable of provoking or inducing an immune response, as with an antigen or a vaccine.
- immunohistochemistry (IHC)
- immunostaining
- The use of an antibody conjugated to a chromophore or fluorophore to bind a specific antigen within a target substance (e.g. a protein) and thereby make the substance visible amidst a background of non-specific substances, allowing for detection of the target in a biological sample. The term originally referred to antibody-based staining of tissue sections with strong dyes or colorants, known as immunohistochemistry, but in modern usage encompasses a much broader range of laboratory methods united by their use of antibodies to label specific biomolecules with visually conspicuous compounds.
- in silico
- (of a scientific experiment or research) Conducted, produced, or analyzed by means of computer modeling or simulation, as opposed to a real-world trial.
- in situ
- (of a scientific experiment or biological process) Occurring or made to occur in a natural, uncontrolled setting, or in the natural or original position or place, as opposed to in a foreign cell or tissue type or in an artificial environment.
- in situ hybridization (ISH)
- A hybridization probe assay in which a labeled, single-stranded DNA or RNA molecule or nucleic acid analogue containing a sequence that is complementary to a particular DNA or RNA sequence is allowed to hybridize with this complement in situ, i.e. in its natural context, such as within cells or tissue sections (as opposed to within homogeneous samples collected from cells or tissues, where cellular or histological structure has been lost in the process of obtaining the sample), in order to reveal the precise location of the particular sequence within this context. The label may be a radioactive compound, fluorescent molecule, or hapten, permitting detection by a variety of visualization techniques. In situ hybridization is commonly used to identify the physical locations of specific DNA sequences such as genes and regulatory elements on chromosomes, which can provide insight into chromosomal structure and integrity; to determine the subcellular locations where various types of RNA accumulate and interact with other molecules; and to visualize the tissues and organs within an organism where specific genes are expressed at various developmental stages (by probing for the genes' RNA transcripts).
- in vitro
- (of a scientific experiment or biological process) Occurring or made to occur in a laboratory vessel or other controlled artificial environment, e.g. in a test tube or a petri dish, as opposed to inside a living organism or in a natural setting.
- in vivo
- (of a scientific experiment or biological process) Occurring or made to occur inside the cells or tissues of a living organism; or, in the broadest sense, in any natural, unmanipulated setting. Contrast ex vivo and in vitro.
- indel
- A term referring to either an insertion or a deletion of one or more bases in a nucleic acid sequence.
- inducer
- A protein that binds to a repressor (to disable it) or to an activator (to enable it).
- inducible gene
- A gene whose expression is either responsive to environmental change or dependent on its host cell's position within the cell cycle.
- in-frame
- 1. (of a gene or sequence) Read or transcribed in the same reading frame as another gene or sequence; not requiring a shift in reading frame to be intelligible or to result in a functional peptide.
- 2. (of a mutation) Not causing a frameshift.[4]
- inheritance
- See heredity.
- initiation codon
- See start codon.
- inosine
- insertion
- A type of mutation in which one or more bases are added to a nucleic acid sequence. Contrast deletion.
- insertion sequence (IS)
- Any nucleotide sequence that is inserted naturally or artificially into another sequence. The term is used in particular to refer to the part of a transposable element that codes for those proteins directly involved in the transposition process, e.g. the transposase enzyme. The coding region in a transposable insertion sequence is usually flanked by short inverted repeats, and the structure of larger transposable elements may include a pair of flanking insertion sequences which are themselves inverted.
- insertional mutagenesis
- The alteration of a DNA sequence by the insertion of one or more nucleotides into the sequence, either naturally or artificially. Depending on the precise location of the insertion within the target sequence, insertions may partially or totally inactivate or even upregulate a gene product or biochemical pathway, or they may be neutral, leading to no substantive changes at all. Many genetic engineering techniques rely on the insertion of exogenous genetic material into host cells in order to study gene function and expression.[4]
- insulator
- A specific DNA sequence that prevents a gene from being influenced by the activation or repression of nearby genes.
- integral membrane protein (IMP)
- Any of a class of membrane proteins which are permanently embedded within or attached to the cell membrane (as opposed to those which are attached only temporarily). Integral membrane proteins can be subclassified into integral polytopic proteins, which span the entirety of the membrane, and integral monotopic proteins, which adhere only to one side.
- integral monotopic protein
- Any of a class of integral membrane proteins which are permanently attached to one side of the cell membrane by any means but which do not completely span the membrane. Contrast integral polytopic protein.
- integral polytopic protein
- Any of a class of integral membrane proteins which span the entirety of the cell membrane, extending from the interior or cytosolic side of the membrane to the exterior or extracellular side. Transmembrane proteins typically have hydrophilic domains exposed to each side as well as one or more hydrophobic domains crossing the nonpolar space inside the lipid bilayer, by which they are further classified as single-pass or multipass membrane proteins. As such many transmembrane proteins function as gated channels or transporters to permit or prohibit the movement of specific molecules or ions across the membrane, often undergoing conformational changes in the process, or as receptors in cell signaling pathways. Contrast integral monotopic protein.
- integration
- integron
- A mobile genetic element consisting of a gene cassette containing the gene for a site-specific recombinase, integrase-specific recognition sites, and a promoter that governs the expression of one or more genes conferring adaptive traits on the host cell. Integrons usually exist in the form of circular episomal DNA fragments, through which they facilitate the rapid adaptation of bacteria by enabling horizontal gene transfer of antibiotic resistance genes between different bacterial species.[4]
- intein
- Any sequence of one or more amino acids within a precursor polypeptide that is excised by protein splicing during post-translational modification and is therefore absent from the mature protein, analogous to the introns spliced out of RNA transcripts.[4] Contrast extein.
- intercalating agent
- Any chemical compound (e.g. ethidium bromide) that disrupts the alignment and pairing of bases in the complementary strands of a DNA molecule by inserting itself between the bases.[2]
- intercalation
- The insertion, naturally or artificially, of chemical compounds between the planar bases of a DNA molecule, which generally disrupts the hydrogen bonding necessary for base pairing.
- intercellular
- Between two or more cells. Contrast intracellular; see also extracellular.
- intercistronic region
- Any DNA sequence that is located between the stop codon of one gene and the start codon of the following gene in a polycistronic transcription unit.[4] See also intergenic region.
- intercross
- A cross in which both the male and female parents are heterozygous at a particular locus.[4]
- intergenic region (IGR)
- Any sequence of non-coding DNA that is located between functional genes.
- intergenic spacer (IGS)
- See spacer.
- interkinesis
- The abbreviated pause in activities related to cell division that occurs during meiosis in some species, between the first and second meiotic divisions (i.e. meiosis I and meiosis II). No DNA replication occurs during interkinesis, unlike during the normal interphase that precedes meiosis I and mitosis.[4]
- internal ribosome entry site (IRES)
- A sequence present in some messenger RNAs that permits recognition by the ribosome and thus the initiation of translation even in the absence of a 5' cap, which in eukaryotes is otherwise required for assembly of the initiation complex. IRES elements are often located in the 5' untranslated region, but may also be found in other positions.
- interphase
- All stages of the cell cycle excluding cell division. A typical cell spends most of its life in interphase, during which it conducts everyday metabolic activities as well as the complete replication of its genome in preparation for mitosis or meiosis.
- intracellular
- Within a cell or cells; i.e. inside the plasma membrane. Contrast intercellular and extracellular.
- intragenic region
- See intron.
- intragenic suppression
- intragenomic conflict
- intrinsic membrane protein
- See integral membrane protein.
- intrinsically disordered protein (IDP)
- intron
- Any nucleotide sequence within a functional gene that is removed by RNA splicing during post-transcriptional modification of the mRNA primary transcript and is therefore absent from the final mature mRNA. The term refers to both the sequence as it exists within a DNA molecule and to the corresponding sequence in RNA transcripts. Contrast exon.
- intron intrusion
- intron-mediated recombination
- See exon shuffling.
- intronic gene
- A gene whose DNA sequence is nested within an intron of another gene and hence surrounded by non-coding intronic sequences.[14]
- invagination
- The infolding of a membrane toward the interior of a cell or organelle, or of a sheet of cells toward the interior of a developing embryo, tissue, or organ, forming a distinct membrane-lined pocket. In the case of individual cells, the invaginated pocket may proceed to separate from the source membrane entirely, creating a membrane-bound vesicle within the cell, as in endocytosis.[8]
- inverted repeat
- A nucleotide sequence followed downstream on the same strand by its own reverse complement. The initial sequence and the reverse complement may be separated by any number of nucleotides, or may be immediately adjacent to each other; in the latter case, the composite sequence is also called a palindromic sequence. Inverted repeats are self-complementary by definition, a property which involves them in many biological functions and dysfunctions. Contrast direct repeat.
- ion channel
- A type of transmembrane protein complex which forms an aqueous pore or channel spanning the lipid bilayer of a membrane, through which specific inorganic, electrically charged ions can diffuse down their electrochemical gradients.[6]
- ionophore
- Any chemical compound or macromolecule that facilitates the movement of ions across biological membranes, or more specifically, any chemical species that reversibly binds electrically charged atoms or molecules. Many ionophores are lipid-soluble proteins that catalyze the transport of monovalent and divalent cations across the hydrophobic lipid bilayers surrounding cells and vesicles.[8]
- isochore
- A large region of genomic DNA with a relatively homogeneous composition of base pairs, distinguished from other regions by the proportion of pairs that are G-C or A-T. The genomes of most plants and vertebrates are composed of different classes of GC-rich and AT-rich isochores.[4]
- isochromosome
- A type of abnormal chromosome in which the arms of the chromosome are mirror images of each other. Isochromosome formation is equivalent to simultaneous duplication and deletion events such that two copies of either the long arm or the short arm comprise the resulting chromosome.
- isomerase
- Any of a class of enzymes which catalyze the conversion of a molecule from one isomer to another, such that the product of the reaction has the same molecular formula as the original substrate but differs in the connectivity or spatial arrangement of its atoms.
- isomeric genes
- Two or more genes that are equivalent and redundant in the sense that, despite coding for distinct gene products, they each result in the same phenotype when set within the same genetic background. If several isomeric genes are present in a single genotype they may be either cumulative or non-cumulative in their contributions to the phenotype.[14]
- isotonic
- Describing a solution containing the same concentration of dissolved solutes as another solution, such that the two solutions have equal osmotic pressure. Isotonic solutions separated from each other by a semipermeable membrane (as with a cell, where the intracellular cytosol is separated from the extracellular fluid by the plasma membrane) have no concentration gradient and thus will not exchange solvent by osmosis. Contrast hypertonic and hypotonic.
J
[edit]- jumping gene
- See transposable element.
- junctional diversity
- junk DNA
- Any DNA sequence that appears to have no known biological function, or which acts in a way that has no positive or a net negative effect on the fitness of the genome in which it is located. The term was once more broadly used to refer to all non-coding DNA, though much of this was later discovered to have a function; in modern usage it typically refers to broken or vestigial sequences and selfish genetic elements, including introns, pseudogenes, intergenic DNA, and fragments of transposons and retroviruses, which together constitute a large proportion of the genomes of most eukaryotes. Despite not contributing productively to the host organism, these sequences are able to persist indefinitely inside genomes because the disadvantages of continuing to copy them are too small to be acted upon by natural selection.
- junk RNA
- Any RNA-encoded sequence, especially a transcript, that appears to have no known biological function, or whose function has no positive or a net negative effect on the fitness of the genome from which it is transcribed. Despite remaining untranslated, many non-coding RNAs still serve important functions, whereas junk RNAs are truly useless: often they are the product of accidental transcription of a junk DNA sequence, or they may result from post-transcriptional processing of primary transcripts, as with spliced-out introns. Junk RNA is usually quickly degraded by ribonucleases and other cytoplasmic enzymes.
K
[edit]- karyogram
- A karyotype which depicts the entire set of chromosomes in a cell or organism by using photomicrographs of the actual chromosomes as they appear in vivo (usually during metaphase, in their most condensed forms), as opposed to the idealized illustrations of chromosomes used in idiograms. The photomicrographs are often still arranged in pairs and by size for easier identification of particular chromosomes, whereas in the actual nucleus there is seldom any apparent organization.
- karyokinesis
- See mitosis.
- karyolymph
- See nucleosol.
- karyon
- See nucleus.
- karyoplasm
- See nucleoplasm.
- karyopyknosis
- See pyknosis.
- karyorrhexis
- The fragmentation and degeneration of the nucleus of a dying cell, during which the nuclear envelope is destroyed and the contents of the nucleus, including chromatin, are dispersed throughout the cytoplasm and degraded by enzymes. Karyorrhexis is usually preceded by pyknosis and may occur as a result of apoptosis, cellular senescence, or necrosis.
- karyosome
- A dense, organized bundle of chromatin which forms in the oocyte nucleus during oogenesis in some female eukaryotes.[26]
- karyotype
- The number and appearance of chromosomes within the nucleus of a eukaryotic cell, especially as depicted in an organized karyogram or idiogram (in pairs and arranged by size and by position of the centromere). The term is also used to refer to the complete set of chromosomes in a species or individual organism or to any test that detects this complement or measures the chromosome number.
- ketogenesis
- The production of ketone bodies via the metabolic decomposition of fatty acids or ketogenic amino acids, an exergonic process which is used to supply energy to certain tissues during periods of carbohydrate and protein insufficiency.
- ketogenic amino acid
- Any amino acid that can be converted directly into acetyl-CoA, which can then be oxidized for energy or used as a precursor for many compounds containing ketone groups. This is in contrast to the glucogenic amino acids, which can be converted into glucose. In humans, seven of the 20 amino acids are ketogenic, though only leucine and lysine are exclusively ketogenic; the other five (phenylalanine, isoleucine, threonine, tryptophan, and tyrosine) are both ketogenic and glucogenic.
- ketolysis
- The metabolic decomposition of ketone bodies, which releases energy that can be used to synthesize ATP.
- kilobase (kb)
- A unit of nucleic acid length equal to one thousand (1×103) bases in single-stranded molecules or one thousand base pairs in duplex molecules such as double-stranded DNA.
- kinase
- Any of a class of enzymes which catalyze the transfer of phosphate groups from high-energy, phosphate-donating molecules such as ATP to one or more specific substrates, a process known as phosphorylation. The opposite process, known as dephosphorylation, is catalyzed by phosphatase enzymes.
- kinesis
- A non-specific, non-directional movement or change in activity by a cell or a population of cells in response to a stimulus, such that the rate of the movement or activity is dependent on the intensity of the stimulus but not on the direction from which the stimulus occurs. Kinesis refers particularly to cellular locomotion without directional bias, in contrast to taxis and tropism.
- kinetochore
- A disc-shaped protein complex which assembles around the centromere of a chromosome during prometaphase of mitosis and meiosis, where it functions as the attachment point for microtubules of the spindle apparatus.
- knob
- In cytogenetics, an enlarged, heavily staining chromomere that can be used as a visual marker, allowing specific chromosomes to be easily identified in the nucleus.[4]
- knockdown (KD)
- A genetic engineering method by which the normal rate of expression of one or more of an organism's genes is reduced or suppressed (though not necessarily completely turned off, as in knockout), either through direct modification of a DNA sequence or through treatment with a reagent such as a short DNA or RNA oligonucleotide with a sequence complementary to either an mRNA transcript or a gene.
- knockin (KI)
- A genetic engineering method in which one or more novel genes are inserted into an organism's genome, particularly when targeted to a specific locus, or in which one or more existing genes are replaced by or substituted with novel genes.[27] This is in contrast to a knockout, in which a gene is deleted or completely inactivated.
- knockout (KO)
- A genetic engineering method in which one or more specific genes are inactivated or entirely removed from an organism's genome, by any of a variety of mechanisms which disrupt their expression at some point in the pathway that produces their gene products, such that no functional gene products are produced. This allows researchers to study the function of a gene in vivo, by observing how the organism's phenotype changes when deprived of the gene's normal effects. A complete knockout permanently inactivates the gene; a conditional knockout allows the gene to be turned on or off at will, e.g. at specific times or in specific tissues, by linking the expression of the gene to some easily modifiable biochemical state or condition. In a heterozygous knockout, only one of a diploid organism's two alleles is knocked out; in a homozygous knockout, both copies are knocked out. Contrast knockin.
- Kozak consensus sequence
- A highly conserved nucleic acid sequence motif which functions as the recognition site for the initiation of translation in most eukaryotic messenger RNAs, generally a sequence of 10 bases immediately surrounding and inclusive of the start codon: GCCRCCAUGG. As the pre-initiation complex scans the transcript, recognition of this sequence (or a close variant) causes the complex to commit to full ribosome assembly and the start of translation. The Kozak sequence is distinct from other recognition sequences relevant to translation such as ribosome binding sites and internal ribosome entry sites.[28]
- Krebs cycle
- See citric acid cycle.

L
[edit]- labelling
- The chemical attachment of a highly selective substance, known as a label, tag, or probe, to a particular cell, protein, amino acid, or other molecule of interest, either naturally or artificially, in vivo or in vitro. Natural labelling is a primary mechanism by which biomolecules specifically identify and interact with other biomolecules; important examples include methylation, acetylation, phosphorylation, and glycosylation. Labelling is also a common laboratory technique, where the label is typically a reactive derivative of a naturally fluorescent compound (e.g. green fluorescent protein), dye, enzyme, antibody, radioactive molecule, or any other substance that makes its target distinguishable in some way. The labelled targets are thereby rendered distinct from their unlabelled surroundings, allowing them to be detected, identified, quantified, or isolated for further study.
- lagging strand
- In DNA replication, the nascent strand for which DNA polymerase's direction of synthesis is away from the replication fork, which necessitates a complex and discontinuous process in contrast to the streamlined, continuous synthesis of the other nascent strand, known as the leading strand, which occurs simultaneously. Because DNA polymerase works only in the 5' to 3' direction, but the lagging strand's overall direction of chain elongation must ultimately be the opposite (i.e. 3' to 5', toward the replication fork), elongation must occur by an indirect mechanism in which a primase enzyme synthesizes short RNA primers complementary to the template DNA, and DNA polymerase then extends the primed segments into short chains of nucleotides known as Okazaki fragments. The RNA primers are then removed and replaced with DNA, and the Okazaki fragments are joined by DNA ligase.
- lampbrush chromosome
- A transcriptionally active, highly de-condensed morphology assumed by certain chromosomes during the diplotene stage of meiotic prophase I in the progenitor cells of oocytes in female insects, amphibians, birds, and some other animals. Lampbrush chromosomes are conspicuous under the microscope because the post-synaptic homologs, still attached at chiasmata, are gigantically elongated into large loops of unpackaged euchromatin extending laterally from a series of chromomeres. Large numbers of messenger RNAs and non-coding RNAs are transcribed from the lateral loops, generating a rich pool of transcripts to be used in the immature oocyte and after fertilization, with functions in both oogenesis and embryogenesis. Because they allow individual transcription units to be directly visualized, lampbrush chromosomes are useful models for studying chromosome organization and genome structure and for constructing high-resolution chromosome maps.[29]
- lateral gene transfer (LGT)
- See horizontal gene transfer.
- leader sequence
- See 5' untranslated region.
- leading strand
- In DNA replication, the nascent strand for which both the direction of synthesis by DNA polymerase and the direction of overall chain elongation are toward the replication fork; i.e. both occur in the 5' to 3' direction, resulting in a single, continuous elongation process with few or no interruptions. By contrast, the other nascent strand, known as the lagging strand, is assembled in a discontinuous process involving the ligation of short DNA fragments synthesized in the opposite direction, away from the replication fork.[4]
- left splicing junction
- The boundary between the left end (by convention, the 5' end) of an intron and the right (3') end of an adjacent exon in a pre-mRNA transcript.
- leptonema
- In meiosis, the first of five substages of prophase I, following interphase and preceding zygonema. During leptonema, the replicated chromosomes condense from diffuse chromatin into long, thin strands that are much more visible within the nucleus.
- lethal mutation
- Any mutation that results in the premature death of the organism carrying it. Recessive lethal mutations are fatal only to homozygotes, whereas dominant lethals are fatal even in heterozygotes.[4]
- leucine zipper (ZIP)
- A common structural motif in DNA-binding transcription factors and some other types of proteins, approximately 35 amino acids in length, characterized chiefly by the recurrence of the amino acid leucine every seven residues. When modeled in an idealized alpha-helical conformation, the leucine residues are positioned in such a way that they can interdigitate with the same or similar motifs in an alpha helix belonging to another similar polypeptide, facilitating dimerization and the formation of a complex resembling a zipper.[12]
- ligand
- In biochemistry, any molecule that binds to or interacts with a specific site on a protein or other biomolecule, usually reversibly via intermolecular forces;[6] or any substance that forms a complex with a biomolecule as part of a biological process. The binding of specific ligands to DNA or proteins is important in many biochemical pathways; for example, protein–ligand binding may result in the protein undergoing a conformational change which alters its function or affinity for other molecules.
- ligase
- A class of enzymes which catalyze the synthesis of large molecules such as nucleic acids by forming one or more chemical bonds between them, typically C–C, C–O, C–S, or C–N bonds via condensation reactions. An example is DNA ligase, which catalyzes the formation of phosphodiester bonds between adjacent nucleotides on the same strand of a DNA molecule, a reaction known as ligation.
- ligation
- The joining of consecutive nucleotides in the same strand of a nucleic acid molecule via the formation of a phosphodiester bond between the 5'-phosphoryl terminus of one nucleotide and the 3'-hydroxyl terminus of an adjacent nucleotide, a condensation reaction catalyzed by enzymes known as ligases.[8] This reaction occurs in fundamentally the same way in all varieties of DNA and RNA catabolism, natural or artificial, whether the addition of individual nucleotides to a growing strand (as in DNA replication and transcription), or the repair of nicks and cuts in previously intact molecules, or the joining of separate nucleic acid fragments into a single molecule (as in chromosomal crossover, exon splicing, retroviral transposition, and all other forms of genetic recombination, as well as artificial molecular cloning techniques). Ligation is the opposite of the anabolic reaction wherein phosphodiester bonds are cleaved by nucleases. It also should not be confused with the non-covalent base pairing that can occur between complementary strands; ligation refers to the synthesis of the phosphate backbone which defines an individual chain of nucleotides.
- linkage
- The tendency of DNA sequences which are physically near to each other on the same chromosome to be inherited together during meiosis. Because the physical distance between them is relatively small, the chance that any two nearby parts of a DNA sequence (often loci or genetic markers) will be separated on to different chromatids during chromosomal crossover is statistically very low; such loci are then said to be more linked than loci that are farther apart. Loci that exist on entirely different chromosomes are said to be perfectly unlinked. The standard unit for measuring genetic linkage is the centimorgan (cM).
- linker DNA
- 1. A short, synthetic DNA duplex containing the recognition sequence for a particular restriction enzyme.[4] In molecular cloning, linkers are often deliberately included in recombinant molecules in order to make them easily modifiable by permitting cleavage and insertion of foreign sequences at precise locations. A segment of an engineered plasmid containing many such restriction sites is sometimes called a polylinker.
- 2. A section of chromosomal DNA connecting adjacent nucleosomes by binding to histone H1.[4]
- linking number
- The number of times that the two strands of a circular double-helical DNA molecule cross each other, equivalent to the twisting number (which measures the torsion of the double helix) plus the writhing number (which measures the degree of supercoiling). The linking number of a closed molecule cannot be changed without breaking and rejoining the strands. DNA molecules which are identical except for their linking numbers are known as topological isomers.[4]
- lipid
- Any of a heterogeneous class of organic compounds, including glycerides (fats), waxes, sterols, and some vitamins, united only by their amphipathic or hydrophobic nature and consequently their very low solubility in water.[12] Some lipids such as phospholipids tend to form lamellar structures or micelles in aqueous environments, where they serve as the primary constituents of biological membranes. Others such as fatty acids can be metabolized for energy, have important functions in energy storage, or serve as signaling molecules. Colloquially, the term "lipids" is sometimes used as a synonym for fats, though fats are more correctly considered a subclass of lipids.
- lipid bilayer
- A lamellar structure composed of numerous amphipathic lipid molecules packed together in two back-to-back sheets or layers, with their hydrophobic fatty acid "tails" directed inward and their hydrophilic "heads" exposed on the outer surface. This is the basic structural motif for all biological membranes, including the plasma membrane surrounding all cells as well as the membranes surrounding organelles and vesicles. Though bilayers are sometimes colloquially described as phospholipid bilayers, phospholipids are just one of several classes of membrane lipids which form bilayers; most membranes are actually a fluid, heterogeneous mixture of phospholipids, glycolipids, and cholesterols, interspersed and studded with various other molecules such as integral proteins.[12]
- lipophilic
- See hydrophobic.
- lncRNA
- See long non-coding RNA.
- locus
- A specific, fixed position on a chromosome where a particular gene or genetic marker resides.
- long arm
- In condensed chromosomes where the positioning of the centromere creates two segments or "arms" of unequal length, the longer of the two arms of a chromatid. Contrast short arm.
- long interspersed nuclear element (LINE)
- Any of a large family of non-LTR retrotransposons which together comprises one of the most widespread mobile genetic elements in eukaryotic genomes. Each LINE insertion is on average about 7,000 base pairs in length.
- long non-coding RNA (lncRNA)
- A class of non-coding RNA consisting of all transcripts of more than 200 nucleotides in length that are not translated. This limit distinguishes lncRNA from the numerous smaller non-coding RNAs such as microRNA. See also long intervening non-coding RNA.
- lyonization
- See X-inactivation.
- lysis
- The disruption and decomposition of the plasma membrane surrounding a cell, or more generally of any membrane-bound organelle or vesicle, especially by osmotic, enzymatic, or other chemical or mechanical processes which compromise the membrane's integrity and thereby cause the unobstructed interchange of the contents of intracellular and extracellular spaces. Lysis generally implies the complete and irreversible loss of intracellular organization as a result of the release of the cell's internal components and the dilution of the cytosol, and therefore the death of the cell. Such a cell is said to be lysed, and a fluid containing the contents of lysed cells (usually including nucleic acids, proteins, and many other organic molecules) is called a lysate. Lysis may occur both naturally and artificially, and is a normal part of the cellular life cycle.
- lysosome
See also
[edit]- Introduction to genetics
- Outline of genetics
- Outline of cell biology
- Glossary of biology
- Glossary of chemistry
- Glossary of evolutionary biology
References
[edit]- ^ "Talking Glossary of Genomic and Genetic Terms". genome.gov. 8 October 2017. Retrieved 8 October 2017.
- ^ a b c d e Klug, William S.; Cummings, Michael R. (1986). Concepts of Genetics (2nd ed.). Glenview, Ill.: Scott, Foresman and Company. ISBN 0-673-18680-6.
- ^ a b Ussery, David W. "DNA Structure: A-, B-, and Z-DNA Helix Families". Lyngby, Denmark: Danish Technical University.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag ah ai aj ak al am King, Robert C.; Stansfield, William D.; Mulligan, Pamela K. (2006). A Dictionary of Genetics (7th ed.). Oxford: Oxford University Press. ISBN 978-0-19-530762-7.
- ^ "NCI Dictionary of Genetics Terms". National Cancer Institute. PDQ® Cancer Genetics Editorial Board.
- ^ a b c d e f g h i j k l m n o p q r Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Raff, Martin; Roberts, Keith; Walter, Peter (2002). "Glossary". Molecular Biology of the Cell (Available from the National Center for Biotechnology Information) (4th ed.). New York: Garland Science.
- ^ a b Lewin, Benjamin (2003). Genes VIII. Upper Saddle River, NJ: Pearson Prentice Hall. ISBN 0-13-143981-2.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae MacLean, Norman (1987). Dictionary of Genetics & Cell Biology. New York: New York University Press. ISBN 0-8147-5438-4.
- ^ López D, Vlamakis H, Kolter R (July 2010). "Biofilms". Cold Spring Harbor Perspectives in Biology. 2 (7): a000398. doi:10.1101/cshperspect.a000398. PMC 2890205. PMID 20519345.
- ^ Momeni B (June 2018). "Division of Labor: How Microbes Split Their Responsibility". Current Biology. 28 (12): R697–R699. doi:10.1016/j.cub.2018.05.024. PMID 29920261. S2CID 49315067.
- ^ "NCI Dictionary of Cancer Terms". www.cancer.gov. National Cancer Institute. 2 February 2011.
- ^ a b c d e f g h i j k l m n o p q r s Lackie, J. M. (2013). The Dictionary of Cell and Molecular Biology (5th ed.). Amsterdam: Academic Press/Elsevier. ISBN 978-0-12-384931-1.
- ^ SwissBioPics. "Animal cell". www.swissbiopics.org. Swiss Institute of Bioinformatics (SIB).
- ^ a b c d e f g h i j k l Rieger, Rigomar (1991). Glossary of Genetics: Classical and Molecular (5th ed.). Berlin: Springer-Verlag. ISBN 3540520546.
- ^ Maheshri N, O'Shea EK (2007). "Living with noisy genes: how cells function reliably with inherent variability in gene expression". Annu. Rev. Biophys. Biomol. Struct. 36: 413–434. doi:10.1146/annurev.biophys.36.040306.132705. PMID 17477840.
- ^ Nishikawa, S. (2007). "Reprogramming by the numbers". Nature Biotechnology. 25 (8): 877–878. doi:10.1038/nbt0807-877. PMID 17687365. S2CID 39773318.
- ^ Connerly, P. L. (2010). "How Do Proteins Move Through the Golgi Apparatus?". Nature Education. 3 (9): 60.
- ^ Valet, Günter (2005). "Cytomics: An entry to biomedical cell systems biology". Cytometry Part A. 63A (2): 67–68. doi:10.1002/cyto.a.20110. ISSN 1552-4922. PMID 15657925.
- ^ Jiao, yan; Katherine Lawler (19 October 2011). "DART: Denoising Algorithm based on Relevance network Topology improves molecular pathway activity inference". BMC Bioinformatics. 12: 403. doi:10.1186/1471-2105-12-403. PMC 3228554. PMID 22011170.
- ^ Priness, I.; Maimon, O.; Ben-Gal, I. (2007). "Evaluation of gene-expression clustering via mutual information distance measure". BMC Bioinformatics. 8: 111. doi:10.1186/1471-2105-8-111. PMC 1858704. PMID 17397530.
- ^ Alberts B, Bray D, Hopin K, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2004). "Tissues and Cancer". Essential cell biology. New York and London: Garland Science. ISBN 978-0-8153-3481-1.
- ^ "Functional Genomics Data Society – FGED Society".
- ^ Edgar, R; Domrachev, M; Lash, AE (1 January 2002). "Gene Expression Omnibus: NCBI gene expression and hybridization array data repository". Nucleic Acids Research. 30 (1): 207–10. doi:10.1093/nar/30.1.207. PMC 99122. PMID 11752295.
- ^ Barrett, T; Wilhite, SE; Ledoux, P; Evangelista, C; Kim, IF; Tomashevsky, M; Marshall, KA; Phillippy, KH; Sherman, PM; Holko, M; Yefanov, A; Lee, H; Zhang, N; Robertson, CL; Serova, N; Davis, S; Soboleva, A (January 2013). "NCBI GEO: archive for functional genomics data sets--update". Nucleic Acids Research. 41 (Database issue): D991-5. doi:10.1093/nar/gks1193. PMC 3531084. PMID 23193258.
- ^ Yakovchuk P, Protozanova E, Frank-Kamenetskii MD (2006). "Base-stacking and base-pairing contributions into thermal stability of the DNA double helix". Nucleic Acids Res. 34 (2): 564–74. doi:10.1093/nar/gkj454. PMC 1360284. PMID 16449200.
- ^ Bogolyubov DS (2018-01-01). Galluzzi L (ed.). "Karyosphere (Karyosome): A Peculiar Structure of the Oocyte Nucleus". International Review of Cell and Molecular Biology. 337. Academic Press: 1–48. doi:10.1016/bs.ircmb.2017.12.001. ISBN 9780128151952. PMID 29551157.
- ^ Gibson, Greg (2009). A Primer Of Genome Science 3rd ed. Sunderland, Massachusetts: Sinauer. pp. 301–302. ISBN 978-0-87893-236-8.
- ^ Kozak, M. (February 1989). "The scanning model for translation: an update". The Journal of Cell Biology. 108 (2): 229–241. doi:10.1083/jcb.108.2.229. ISSN 0021-9525. PMC 2115416. PMID 2645293.
- ^ Morgan, G.T. (2002) "Lampbrush chromosomes and associated bodies: new insights into principles of nuclear structure and function." Chromosome Research. 10: 177–200.
Further reading
[edit]- Budd, A. (2012). "Introduction to Genome Biology: Features, Processes, and Structures". Evolutionary Genomics. Methods in Molecular Biology. Vol. 855. pp. 3–4. doi:10.1007/978-1-61779-582-4_1. ISBN 978-1-61779-581-7. PMID 22407704.
